ISSN: 0973-7510
E-ISSN: 2581-690X
MRSA causes nosocomial and community based infections. It is associated with significant mortality and morbidity. Resistance in MRSA is encoded by mecA gene. Anterior nares are the ecological niche of Staphylococcus aureus. HCWs who are colonized with MRSA, act as agents of cross contamination of hospital and community acquired MRSA. Treating MRSA infections is a therapeutic challenge as it is resistant to beta lactam group of drugs. Therefore, there is a need for rapid and accurate detection of MRSA carriage in HCWs and to understand its antibiotic susceptibility pattern.The objective of the present study is to estimate the occurrence of MRSA in HCWs, using phenotypic and genotypic methods. A prospective study for six months was conducted after obtaining Institutional Ethical Committee clearance. Anterior nasal swabs of those HCWs who gave informed consent were taken processed for culture and sensitivity as per standard protocol. To detect MIC for oxacillin, E-strip method was used. mecA gene detection was done by PCR. A total of 300 HCWs were sampled.14.66% (44/300) of the isolates were identified as Staphylococcus aureus, of which 10 isolates were detected as MRSA. The overall isolation rate of MRSA is 3.33 %(10/300). MRSA carriage was high amongst nurses (5/59, 8.47%), followed by doctors (4/105, 3.80%).Antibiotic sensitivity pattern showed that highest resistance was to penicillin (75%) followed by amoxiclav (70.45 %).9 MRSA isolates were detected as mecA gene positive by PCR. MRSA carriers were decontaminated successfully with 2% mupirocin ointment and 2% chlorhexidine shampoo. This study reiterates the need for rapid and accurate identification of HCWs who have nasal colonization with MRSA, for reinforcing hospital infection control measures and decontamination protocol. This will help prevent the spread of MRSA in our community.
Decolonization, E-strip, Health care workers, mecA gene, Methicillin resistance, nasal carriage of MRSA
Staphylococcus aureus is an established pathogen, which can cause a broad spectrum of infection as it has many virulence factors. It has been observed that 50% of the adult population is either persistent or intermittent carriers of Methicillin resistant Staphylococcus aureus (MRSA).1One of the commonest cause of community based and nosocomial infections is MRSA. It is known to cause significant mortality and morbidity. One of the challenges that clinicians face with infections caused by MRSA is that there are limited therapeutic options, as resistance to beta lactam antibiotic, means that the organism is resistant to beta lactam group of drugs.2
Resistance in MRSA is due to the presence of Penicillin binding protein (PBP2a) which is encoded by mecA gene. The mecA gene is present on the Staphylococcal Cassette Chromosome (SCC). Detection of the mecA gene by PCR is rapid, accurate and is considered the gold standard. 3
Infections caused by MRSA are associated with prolonged hospital stay, higher cost of treatment and worst outcomes. 4Several studies have observed that the anterior nares are the ecological niche of Staphylococcus aureus and that these cause invasive infections.4, 5The main sources of MRSA in a hospital setting are the asymptomatically colonized patients and HCWs. 2, 6, 7Health care workers (HCWs) form the interface between the hospital and the community. HCWs can act as agents of cross contamination of hospital acquired and community acquired MRSA. 8The reason for this being, poor compliance to hand hygiene by the HCWs. 9, 10 The rate of MRSA nasal carriage in HCWs ranges from 6.0% to 17.8 %. 9
Early identification of HCWs colonized with MRSA combined with appropriate hand hygiene and other precautions can control the spread of MRSA.11Therefore, there is a need for rapid and accurate detection of MRSA carriage in HCWs and the susceptibility pattern of the isolated MRSA strain. This will help the hospital to strengthen its infection control practices and initiate decolonization in HCWs who are MRSA positive. As a result, the spread of MRSA can be controlled. Hence, we undertook this study to detect nasal carriage rate of MRSA in our HCWs, rapidly and accurately. We also looked in to the antibiotic susceptibility pattern of the isolates, so that we could initiate decolonization in such HCWs and strengthen the infection control practices in our teaching hospital.
Objectives
- To estimate the occurrence of MRSA in HCWs working in our teaching hospital using phenotypic and genotypic methods.
- To understand the antibiotic susceptibility pattern of the isolated Staphylococcus aureus.
- To initiate appropriate decolonization and infection control measures.
The study protocol was submitted to the Institutional Ethical Committee and necessary permission was obtained to conduct this study. A prospective study for a period of six months was conducted in our teaching hospital. Informed consent was obtained from HCWs before taking nasal swabs. Only those HCWs who gave informed consent were included in this study. Those HCWs who suffered from fever, upper respiratory tract infections, who had a history of recent nasal surgery and who were on topical nasal medication were excluded from this study. 2
The swabs were inoculated on to Mannitol salt agar, blood agar, Mac Conkey agar and BHI broth (HiMedia, Mumbai).
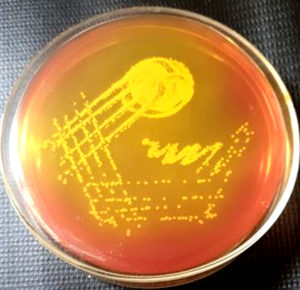
These were incubated at 37ºC for 24 hours. Methicillin resistant Staphylococcus aureus gives yellow colonies on mannitol salt agar. These yellow colonies were further processed as per standard protocol (Gram stain, catalase test, urease test, anaerobic fermentation of mannitol and tube coagulase test) to identify them as Staphylococcus aureus. 12, 13
Antibiotic sensitivity testing
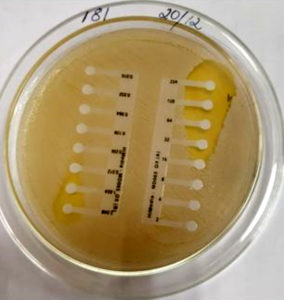
The antibiotic sensitivity testing was done using Kirby Bauer disc diffusion method. The following antibiotic were used: amoxyclav (30 µg /10 µg), cefoxitin (30 µg), chloramphenicol (30 µg), ciprofloxacin (5 µg), clindamycin (2 µg), cotrimoxazole ( 1.25/23.75 µg), doxycycline (30 µg), erythromycin (15 µg), gentamicin (10 µg), linezolid ( 30 µg),oxacillin(1 µg ) and penicillin ( 10 units), vancomycin(30 µg ) and teicoplanin(30 µg). The interpretation of zone diameters into sensitive, intermediate, and resistant was done as per the Clinical and Laboratory Standards 2019. 14, 15
The following methods were used to detect methicillin resistance
Oxacillin disc diffusion
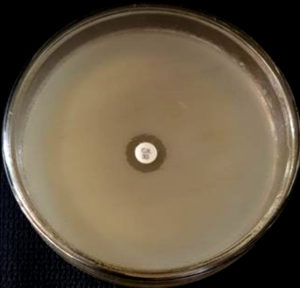
The Staphylococcus aureus isolates were inoculated on Mueller Hinton agar plates (HiMedia, Mumbai) with 4% NaCl. The inoculum was standardized to 0.5 McFarland units. An oxacillin disc of 1 µg (HiMedia, Mumbai) was placed on the inoculated plate. The inoculated plates were incubated at 350C for 16-29 hours. A zone diameter lesser than or equal to 10 mm is considered as resistant to oxacillin (MRSA positive), as per the CLSI guidelines. Quality control was done using ATCC strain Staphylococcus aureus 25923. 16, 17
Cefoxitin disc diffusion
The Staphylococcus aureus isolates were inoculated on Mueller Hinton agar plates (HiMedia, Mumbai). The inoculum was standardized to 0.5 McFarland units. The inoculated plates were incubated at 370C for 16-18 hours. To detect MRSA, we used a cefoxitin disc (30 µg, HiMedia, Mumbai). Cefoxitin disc diffusion results are more sensitive and easy to detect when compared to oxacillin disc diffusion. A zone diameter of lesser than or equal to 21 mm is considered resistant to cefoxitin (MRSA positive). Quality control was done using ATCC strain Staphylococcus aureus 25923. 16, 17
E-strip method
MIC for oxacillin was tested using Hi Comb MIC strip (HiMedia, Mumbai) .This test was done on Mueller Hinton (HiMedia, Mumbai) agar plates with 2% NaCl. The inoculum was standardized to 0.5 McFarland units. The MIC applicator was used to place the E- strip on the inoculated plate. The inoculated plates were incubated at 350C for 24 hours. The MIC lesser than or equal to 2 microgram was considered sensitive and and greater than or equal to 4 microgram was considered resistant.17
mecA gene detection by PCR
The 10 MRSA isolates were subjected to a triplex PCR. The targets for the triplex PCR are:
- 16S rRNA (Staphylococcus genus specific),
- nuc(Staphylococcus aureus species specific) and
- 3. mecA (gene for methicillin resistance)
To detect the 16S rRNA, which is genus specific, forward primer Staph756F with the oligonucleotide sequence AACTCTGTTATTAGGGAAGAACA was used. The reverse primer Staph750R with the oligonucleotide sequence CCACCTTCCTCCGGTTTGTCACC was used.
To detect the nuc gene, which is species specific, forward primer Nuc F with the oligonucleotide sequence GCGATTGATGGTGATACGGTT was used. The reverse primer with the oligonucleotide sequence AGCCAAGCCTTGACGAACTAAAGC was used.
To detect the mecA gene, which detects methicillin resistance, forward primer MecA1 F with the oligonucleotide sequence GTAGAAATGACTGAACGTCCGATAA was used. The reverse primer MecA2 R with the oligonucleotide sequence AGCCAAGCCTTGACGAACTAAAGC was used. The composition for a 25 µl reaction mixture for PCR consisted of : PCR master mix 2X(12.5 µl), Staph756F primer – 20 µM(1 µl), Staph750R primer – 20 µM(1 µl), Nuc F primer – 10 µM(1 µl), Nuc R primer – 10 µM(1 µl), MecA1 F primer – 10 µM(1 µl), MecA2 R primer – 10 µM(1 µl), DNA template 100 -200 ng (2 µl)and Nuclease free water(4.5 µl).Thermal cycler conditions for triplex PCR are as follows: 1 cycle of initial denaturation at 94ºC for 5 minutes. The next cycle of denaturation was done at 94ºC for 1 minute, annealing at 600C for 1 minute and extension at 72ºC for 2 minutes (all put together 30 cycles).The final extension was done at 72ºC for 10 minutes (I cycle).A 2% agarose gel containing 10 µg/ml ethidium bromide was used to capture the amplicons. The amplicon sizes for 16S rRNA, nuc gene and mecA gene are 756 bp, 279 bp and 310 bp respectively. Primers from Sigma Company were used. The PCR master mix that was used was from Takara Company, Japan. For the agarose gel electrophoresis, the gel image was captured using gel documentation system (G:BOX, system Syngene).18,19,20
Statistical analysis
Data entry was done using Microsoft Excel sheet. The data was analysed using SPSS version 22. Percentage analysis of the data was given.
A total of 300 health care workers were sampled. The sex distribution among the health care workers who were sampled, is as follows: were 30% were males (90/300) and 70% (210/300) were females. (Table 1)
Table (1):
Sex distribution among the health care workers who were sampled.
Sex |
Number (n=300) |
Percentage (%) |
|---|---|---|
Male |
90 |
30 |
Female |
210 |
70 |
Total |
300 |
100 |
The highest numbers (42%, 126/300) of healthcare workers who were sampled were in the age group of 21-30 years, followed by 31-40 years age group (32.3%, 97/300).15 %( 45/300) of the HCWs belonged to the 41-50years age group. 6.6 %( 20/300) of the HCWs belonged to the ≥ 50 years age group and 4% (12/300) of the HCWs belonged to the 18-20 years age group. (Table 2)
Table (2):
Age distribution among the health care workers who were sampled
Age Group |
Total Number (n=300) |
Percentage (%) |
|---|---|---|
18 – 20 years |
12 |
4.0 |
21 – 30 years |
126 |
42.0 |
31 – 40 years |
97 |
32.3 |
41 – 50 years |
45 |
15.0 |
> 50 years |
20 |
6.6 |
Total |
300 |
100 |
Among the health care workers who were sampled 35% (105/300) were doctors,32%(96/300) were housekeeping staff, 19.6%(59/300) were nurses, 7%(21/300) were technicians and 6.3% (19/300) were office staff .(Table 3)
Table (3):
Distribution of health care workers who were sampled.
Category |
Total Number (n=300) |
Percentage (%) |
|---|---|---|
Doctors |
105 |
35.0 |
Nurses |
59 |
19.6 |
Technicians |
21 |
7.0 |
Office staff |
19 |
6.3 |
House keeping |
96 |
32.0 |
Total |
300 |
100 |
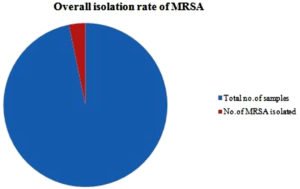
Of the 300 nasal swabs, 14.66% (44/300) of the isolates were identified as Staphylococcus aureus. Of these 44 isolates, only 10 isolates were detected to be MRSA positive. So the isolation rate of MRSA, from the Staphylococcus aureus isolates is 22.72 %( 10/44).The overall isolation rate of MRSA is 3.33 %( 10/300). (Figure 1)
Out of the 300 healthcare workers screened 90 were males and among them 1(1.1%) was an MRSA carrier. Of the 210 females, 9(4.2%) were MRSA carriers. (Table 4)
Table (4):
Gender distribution among the MRSA carriers.
Gender |
Number screened |
No. of MRSA carriers |
Percentage of MRSA carriers (%) |
|---|---|---|---|
Male |
90 |
1 |
1.1 |
Female |
210 |
9 |
4.2 |
In our study, MRSA carriage was high among the nurses (5/59, 8.47%), followed by doctors (4/105, 3.80%) and housekeeping staff (1/96, 1.04%). No MRSA was isolated in the technicians and the office staff. (Table 5)
Table (5):
Isolation rate of MRSA in different professional categories.
Profession |
MRSA number |
MRSA (%) |
|---|---|---|
Doctors (n=105) |
4 |
3.80 |
Nurses(n=59) |
5 |
8.47 |
House Keeping(n=96) |
1 |
1.04 |
Technicians(n=21) |
0 |
0 |
Office staff(n=19) |
0 |
0 |
Of the 300 HCW screened, 4 MRSA carriers were from labour OT (3 female nurses and 1 male nurse).1 female nurse from casualty was detected to be a MRSA carrier. Amongst the doctors, who were detected as MRSA carriers, 2 female doctors were from the Department of Biochemistry, 1 female doctor from the Department of Anaesthesia and 1 female doctor from the Department of Pathology. Amongst the housekeeping staff, 1 female housekeeping staff from special ward was reported to be a MRSA carrier. (Table 6)
Table (6):
Professional category, gender and department wise distribution of MRSA carriers.
Category |
Gender |
Department |
No. of MRSA carriers |
|---|---|---|---|
Doctors |
F |
Anaesthesia |
1 |
Doctors |
F |
Pathology |
1 |
Doctors |
F |
Biochemistry |
2 |
Nurse |
F |
Labour OT |
3 |
Nurse |
M |
Labour OT |
1 |
Nurse |
F |
Casualty |
1 |
House Keeping |
F |
Special ward |
1 |
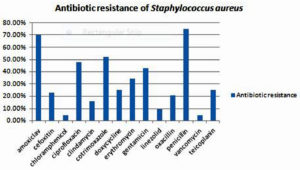
Antibiotic sensitivity pattern of the 44 Staphylococcus aureus isolates, 70.45 %(31/44) were resistant to amoxiclav, 22.73% (10/44) were resistant to cefoxitin,4.55% (2/44) were resistant to chloramphenicol, 47.73% (21/44) were resistant to ciprofloxacin,15.91%(7/44) were resistant to clindamycin,52.27%(23/44) were resistant to cotrimoxazole,25%(11/44) were resistant to doxycycline,34.09%(15/44) were resistant to erythromycin,43.18%(19/44) were resistant to gentamicin,9.09%(4/44) were resistant to linezolid, 20.45%(9/44) were resistant to oxacillin,75%(33/44) were resistant to penicillin, 4.55% (2/44) were resistant to vancomycin and, 25% (11/44) were resistant to teicoplanin.(Figure 5)
The various areas of our hospital, from which Methicillin resistant Staphylococcus aureus(MRSA) were isolated is as follows:4 MRSA carriers were from labour OT and 1 each from Department of Anaesthesia and Pathology, 2 from the Department of Biochemistry and 1 each from Special ward and Casualty.(Table 7)
Table (7):
Isolation rate of MRSA in different studies.
Sl.no. |
Study |
Rate of isolation of MRSA (%) |
|---|---|---|
1. |
Present study |
3.33 |
2. |
Kausalyaet al.,2015 |
1.33 |
3. |
Radhakrishna M et al.,2013 |
17.5 |
4. |
Gargi- Dangre- Mudey et al.,2015 |
2.08 |
5. |
Agarwal L et al.,2015 |
14.0 |
6. |
Nabil Abdullah El Aila et al.,2017 |
25.5 |
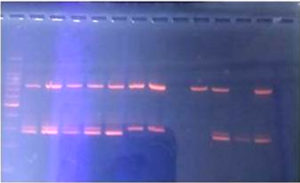
In our study, the PCR detected only one MRSA isolate as mecA gene negative. The remaining 9 MRSA isolates were detected as mecA gene positive. (Figure 6)
Several studies have observed that detection of MRSA carriage among HCWs is important, as they act as potential reservoirs of nosocomial infections. They help in the spread of MRSA not only to patients and inanimate surfaces, but also to other HCWs and their family members.4, 5
In our study, the nasal carriage of Staphylococcus aureus in HCWs is 22.72 %. The nasal carriage of MRSA in HCWs is 3.33%.
The difference in the nasal carriage rate noted in various studies can be attributed to the differences in the sample size, study design and the MRSA detection methods that were used. 4Various studies report different isolation rates.
(Refer Table 7)
In our study, 10 % of the MRSA carrier were males (1/10) and 90% of the MRSA carriers were females (9/10).This is in contrast with other studies where a male preponderance in the MRSA carriers has been reported. 2However, further studies need to be undertaken to ascertain this, as the sample size and number of male and female HCWs were not matched.
In our study, nasal carriage of MRSA was high among nurses (8.47%), followed by doctors (3.80%) and the housekeeping staff (1.04%). This finding is similar to a study done Mondal et al9 in their study , the nasal carriage amongst the nurses was 25% and 18.8% carriage rate in doctors. Several studies have observed that the nasal carriage of MRSA is high among nurses, followed by doctors. 6, 8
In our study, 5 nurses were detected as MRSA carriers. Of these 5 nurses, only one was a male nurse.4 nurses were from the Labour OT and 1 was posted in Casualty. In our study, 4 doctors were detected as MRSA carriers. All the 4 doctors were females.3 doctors were from the Central Lab (1 from Pathology, and 2 from Biochemistry). Several studies have identified HCWs from Central Lab to be carriers of MRSA. Our study is in agreement with them. The reason for this could be that HCWs who are from the Central Lab are exposed to a large number of patients. They also handle large number of infected samples. 5In our study, only 1 female housekeeping staff was detected as a MRSA carrier. She was posted in Special ward.
Antibiotic sensitivity pattern of the Staphylococcus aureus isolates
In our study, the highest resistance was seen to penicillin (75%). Most of the studies report 100% resistance to penicillin. The second highest resistance was observed to amoxyclav (70.45%).With regards to ciprofloxacin, we observed a resistance of 47.73%. This is similar to the ciprofloxacin resistance (50%), reported by Agarwal L et al study. 6 Resistance was observed to cotrimoxazole (52.27%),doxycycline (25.00%),erythromycin (34.09%) and gentamicin (43.18%).
Over the last decade, there are reports of glycopeptide and vancomycin resistance. 5In our study, we observed 4.55% resistance to vancomycin. In a study conducted by El Aila et al., in 2017, reports 15.7% resistance to vancomycin .8 In our study, 2 MRSA isolates showed resistance to vancomycin. These two isolates belonged to two female labour OT nurses. Good sensitivity was seen to chloramphenicol (95.45%), vancomycin (95.45%) linezolid (90.91%) and clindamycin (80.09%).
The multidrug resistance pattern that is observed in this study can be attributed to overuse/misuse/irrational use of antibiotics and poor compliance to hospital antibiotic policy. However these causes need to be thoroughly looked into.
Several studies have proved that cefoxitin disc diffusion correlates better with the presence of mecA gene. In our study, out of the 44 Staphylococcus aureus isolates, 9 isolates (9/44, 20.45%) were detected as MRSA by oxacillin disc diffusion, whereas 10 isolates (10/44, 22.72%) were detected as MRSA by cefoxitin disc diffusion. However, all 10(10/44, 22.72%) isolates detected as MRSA cefoxitin disc diffusion, had a MIC greater than 4 microgram by the E-strip method.
The gold standard for the detection of MRSA is PCR. It detects the mecA gene. The PCR results are available within 24 hours. when compared to traditional culture or phenotypic techniques which take 48 hours. This can help the clinician in initiating accurate and early treatment. Therefore, PCR is preferred over the phenotypic methods.
In our study, out of the 10 MRSA isolates, one isolate was mecA gene negative on PCR. The reason for one isolate to be mecA gene negative on PCR could be:
- mecA gene expression levels could differ for every104 to 106
- due to the absence of the penicillinase plasmid. This plasmid maintains the stability and phenotypic expression of the mecA gene.3
- due to mutations in the primer binding sites, within the gene.8
The one isolate which was mecA gene negative on PCR, belonged to the male nurse, who was from the Labour OT.
It has been observed that HCWs with nasal colonization of MRSA are at a fourfold high risk to get infection. They also transmit multidrug resistant strains not only to other HCWs, patients and family members, but also to the community at large. Therefore, it is important to initiate the decontamination protocol. In our study, we identified 10 HCWs who had nasal colonization with MRSA. All the 10 MRSA carriers were identified and contacted personally by us. We scheduled a time and place in our Microbiology Department to meet each one of them individually. We maintained confidentiality and privacy when we broke the news to them, that they were identified as MRSA carriers. The laboratory reports were shown to them and the same was discussed with them. They were counseled to take up the decontamination protocol. All the 10 MRSA carriers agreed to take up the decontamination process. They were educated and instructed to apply 2% mupirocin ointment, twice daily to each of their nostrils. They were also instructed to shampoo daily, for a week, using 2% chlorhexidine shampoo. 2After this, two sets of cultures were performed with 72 hours apart. All the 10 MRSA carriers were negative, after the completion of the decontamination protocol. The findings of this study were placed and discussed with the Hospital Infection Control Committee of our teaching hospital, to further initiate appropriate infection control measures.
Limitations
All the HCWs could not be swabbed. It is important that all HCWs are screened for MRSA especially the ones working in critical care units. The vancomycin resistance that was observed in this study, could not be confirmed by other phenotypic (agar or broth dilution for MIC) or genotypic methods (PCR). In this study, we could not investigate further, on that one isolate which was detected as mecA gene negative, as it was beyond the scope of this study.
This study reiterates the need for rapid and accurate identification of those HCWs who have nasal colonization with MRSA, to prevent its further spread in the health care setting as well as in the community. This study also presses the need for reinforcing hospital infection control measures like hand hygiene and use of personal protective equipment (mask and gloves).It is also important that all health care settings should implement an ongoing surveillance programme to detect and decontaminate HCWs who are identified as MRSA carriers. This should be combined with awareness programmes conducted for all categories of HCWs. This multi-pronged approach will help prevent the spread of MRSA in our community.
ACKNOWLEDGMENTS
We would like to thank the Managing Director of BGS Group of Institutions Sri Sri Dr. Prakashnath Swamiji for all the encouragement he gives to research. We would like to thank our dynamic Principal Dr. M.E. Mohan Sir, for his guidance. We are very grateful to Dr. Veena and Mrs. Jyothi from NIMHANS, Bangalore for helping us with the PCR. We would like to thank all the HCWs who actively participated in this study. We convey our heartfelt gratitude to the teaching and non-teaching staffs of the Department of Microbiology, who have supported us.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both the authors have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This study was conducted after obtaining necessary approval from the Institutional Ethical Committee; Protocol number BGS GIMS/IEC/1301 dated 23.03.2017.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Pantosti A, Venditti M. What is MRSA? European Respiratory Journal.2009;34:1190-1196.
Crossref - Kausalya, Kashid RA, Sangeetha S. Nasal carriage and antimicrobial susceptibility of Staphylococcus aureus, with special reference to methicillin resistance, in health care workers in tertiary care hospitals in South India. Scholars Academic Journal of Biosciences. 2015; 3(8):720-724.
- Khan SAK, Shetty PJ, Sarayu LY, Chidambaram A, Ranganathan R. Detection of mecA genes of Methicillin-Resistant Staphylococcus aureus by Polymerase Chain Reaction. Int J Health Rehabil Sci. 2012;1(2): 64-68.
Crossref - Radhakrishna M, D’Souza M, Kotigadde S, Vishwas SK, Shashidar KM. Prevalence of Methicillin Resistant Staphylococcus aureuscarriage amongst Health Care Workers of Critical Care Units in Kasturba Medical College Hospital, Mangalore, India. J Clin Diagn Res. 2013;7(12):2697–2700.
- Mudey GD, Mudey A, Tankhiwale N. Study of nasal carriage of Staphylococcus aureusamong health care workers of a rural tertiary health care centre. Int J Healthcare and Biomed Res. 2015;3(3):85-90.
- Agarwal L, Singh AK, Sengupta C, Agarwal A. Nasal carriage of Methicillin- and Mupirocin-resistant S. aureus among health care workers in a tertiary care hospital. Journal of Research in Pharmacy Practice. 2015;4(4):182-186.
Crossref - Mathanraj S, Sujatha S, Sivasangeetha K, Parija SC. Screening for Methicillin-resistant Staphylococcus aureus carriers among patients and health care workers of a tertiary care hospital in South India. Indian J Med Microbiol. 2009;27(1):62-64.
Crossref - Aila NAE, Laham NAA and Ayesh BM. Nasal carriage of methicillin resistant Staphylococcus aureus among health care workers at Al Shifa hospital in Gaza Strip. BMC Infectious Diseases. 2017;17:28.
Crossref - Mondal H, Gupta I, Nandi P, Ghosh P, Chattopadhyay S, Mitra GD. Nasal Screening of Healthcare Workers for Nasal Carriage of Methicillin Resistance Staphylococcus aureus, Vancomycin Resistance Staphylococcus aureus and Prevalence of Nasal Colonization with Staphylococcus aureus in Burdwan Medical College and Hospital. International Journal of Contemporary Medical Research. 2016;3(11):3342-3346.
- JM Boyce. MRSA patients proven methods to treat colonization and infection. J Hosp. Infect. 2001;48:S9-S14.
Crossref - Khanal R, Sah P, Lamichhane P, Lamsal A, Upadhaya S, Pahwa VK. Nasal carriage of methicillin resistant Staphylococcus aureus among health care workers at a tertiary care hospital in Western Nepal. Antimicrobial Resistance and Infection Control. 2015;4:39.
Crossref - Baird D. Staphylococcus: Cluster forming Gram-positive cocci, chapter 11. In: Mackie and McCartney Practical Medical Microbiology, 14th ed. Collee JG, Fraser AG, Marimon BP, Simmons A, editors. Churchill Livingstone: New York; 1996. p.245.
- Winn WC, Allen SD, Janda WM, et al, Gram positive cocci Part I: Staphylococci and related Gram positive cocci, In: Koneman’s colour atlas and textbook of diagnostic Microbiology. 6 th ed. USA: Lippincot Williams and Wilkins; 2006 p. 623 -673.
- Clinical and Laboratory Standard Institute. Performance standards for antimicrobial disc susceptibility tests, twentieth supplement. 2012; 32:M100–S21.
- PA Wayne. Clinical and Laboratory Standard Institute; 2013. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard-Eleventh Edition and Performance Standards for Antimicrobial Susceptibility Testing: Twenty- Third Informational Supplement. CLSI Document M02-A11 and M100-S23.
- Broekema NM, Van TT, Monson TA, Marshall SA, Warshauer DM. Comparison of Cefoxitin and Oxacillin Disk Diffusion Methods for Detection of mecA-Mediated Resistance in Staphylococcus aureus in a Large-Scale Study. Journal of Clinical Microbiology. 2009;217–219.
Crosref - Sharma S, Srivastava P, Kulshrestha A, Abbas A. Evaluation of different phenotypic methods for the detection of methicillin resistant Staphylococcus aureus and antimicrobial susceptibility pattern of MRSA. Int J Community Med Public Health. 2017; 4:3297-301.
Crossref - Jaffe RI, Lane JD, Albury SV , Niemeyer DM. Rapid Extraction from and Direct Identification in Clinical Samples of Methicillin-Resistant Staphylococci Using the PCR. Journal of Clinical Microbiology. 2000; 38(9):407-3412.
Crossref - Al AG-Akydy, Daoud H, Mulhem MM. Disc diffusion method versus PCR for mec A gene in detection of oxacillin resistant Staphylococcus aureus in University Children’s Hospital in Damascus, Syria. International Journal of Pharmacy and Pharmaceutical Sciences. 2014; 6(4)488-491.
- Elhassan MM,Ozbak HA, Hemeg HA, Elmekki MA, Ahmed LM. Absence of the mecA Gene in Methicillin Resistant Staphylococcus aureus isolated from different clinical specimens in Shendi City, Sudan. Biomed Res Int. 2015;2015:895860. PMCID: PMC4531171.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.