Anuj Bansal *and Mohammad Shahid
Biocontrol Laboratory, Department of Plant Pathology, Chandra Shekhar Azad University of Agriculture & Technology, Kanpur – 208 002, India.
ABSTRACT
Most isolates of the genus Trichoderma were found to act as mycoparasites of many economically important aerial and soil-borne plant pathogens. Trichoderma has gained importance as a substitute for chemical pesticides and hence an attempt was intended to corroborate the positive relatedness of molecular and morphological characters. A fungal strain of Trichoderma asperellum CA-03/9840 was isolated from a soil sample collected from Farmer Field, Sultanpur ,Uttar Pradesh, India. The universal primers were used for amplification of 5.8SrRNA gene fragment and the strain was characterized by using 5.8SrRNA gene sequence with the help of ITS marker. It is proposed that the identified strain Trichoderma asperellum CA-03/9840 be assigned as the type strain of a species of the genus Trichoderma based on Tricho Key analysis together with the 5.8SrRNA gene sequence search in Ribosomal Database Project, small subunit rRNA and large subunit rRNA databases. The sequence was deposited in Gene Bank with the accession number KU821782. Thus an integrated approach of morphological and molecular markers can be employed to identify a superior strain of Trichoderma for its commercial exploitation.
Keywords: 5.8S ribosomal RNA gene, Trichoderma, ITS, Biocontrol, Antagonist.
INTRODUCTION
Trichoderma, commonly available in soil and root ecosystem, has gained immense importance since last few decades due to its biocontrol ability against several plant pathogens. Some strains of Trichoderma like T. harzianum, T. atroviride, T. viride, T. virens and T. koningii are efficient bio control agents which have the ability to inhibit pathogen growth in the soil, hence improving the overall health of the plant. Antagonistic microorganisms such as Trichoderma reduce growth, survival of pathogen by different mechanism like competition antibiosis, mycoparasitism, hyphal interactions and enzyme secretion. Such micro organisms are now available commercially and are used in crop management and practices Kubicek, et al. (2000). The use of Trichoderma species as biological control agents has been investigated for over 70 years but it is only relatively recently that strains have become available commercially. Biocontrol agents are widely regarded by public as “natural” and non-threatening products, although risk assessments must clearly be carried out on their effects on non-target organisms. Moreover, knowledge concerning the behaviour of such antagonists is essential for their effective use.The morphological and microscopic figure of Trichoderma asperellum CA-03/9840 is given figure 1 (a-c).
Trichoderma asperellum
- asperellum was described as a new species in 1999 for isolates that produce finely warted conidia, with temperature optima of 300C and survive by producing chlamydospore. It is used as biological control agent against a wide spectrum of plant disease-causing organisms, including straminopiles such as Phytophthora megakarya fungi and nematodes. The species also has been shown to have antibacterial activity through the production of trichotoxin peptaibols.
Habit and Habitat
Isolate Name: Tasp/CA-03
Isolated from: Sultanpur
Field: Chilli
ITCC No.: 9840
Morphological Key:
Morphological Observation |
Species Character |
Colony growth rate (cm) |
7-8 cm |
Colony colour |
Dark yellowish green |
Reverse colony colour |
Colourless |
Colony edge |
Smooth |
Culture smell |
No characteristic odour |
Mycelial form |
Smooth |
Mycelial colour |
Cottony white |
Cultural characteristic:
Cultural Character |
Species Character |
Conidiation |
Ring like zones |
Conidiophore branching |
Branched, regular |
Phialide shape |
Cyllindrical |
Phialide size |
6.2-10.5 x 3.1-3.9 mm |
Conidial shape |
Globose to subglobose |
Philospore size |
2.6-3.0 x 2.0-2.4 mm |
Conidial wall |
Smooth |
Conidial colour |
Light green |
Chlamydospore |
present |
Spore Germination time |
12 -13 Hours |
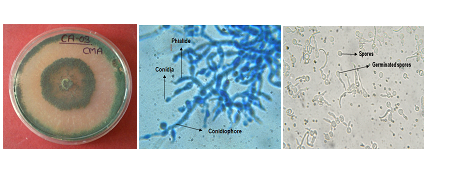
(A) Growth on PDA Media (B) Light micrographs of T. asperellum at 40x (C) Spore germination at 40x
Fig. 1 (A-C): Trichoderma asperellum CA-03/9840 strain in PDA medium and microscopic image
Accurate and definitive fungal identification is essential for correct disease diagnosis, treatment of associated with fungal infections. Characterization of fungal species using classical methods is not as specific as the genotyping methods. Genotypic techniques involve the amplification of a phylogenetically informative target, such as the small-subunit (5.8S) rRNA gene reported by Woese CR et. al. (1977). rRNA is essential for the survival of all cells, and the genes encoding the rRNA are highly conserved in the fungal and other kingdoms. The sequences of the rRNA and proteins comprising the ribosome are highly conserved throughout evolution, because they require complex inter- and intramolecular interactions to maintain the protein-synthesizing machinery reported by Sacchi C. T., et al. (2002), Hillis D. M., et al. (1991) and Woese C. R. et al (1987). Trichoderma species are common soil inhabitants and are effective in providing bio-control of soil borne pathogens due to antagonistic behaviors. The major aspect of successful biological control strategies includes the production, formulation and delivery system of bio-agents.The internal transcribed spacer (ITS) region of the rDNA is perhaps the most widely sequenced DNA region in fungi. It has typically been most useful for molecular systematic study at species level, and even within species found by Ospina-Giraldo M. D. et al. (1998), Kubicek, C. P. et al. (2000), Kulling-Gradinger C. M. et al. (2002) and Lee, C. et al. (2002) attempted a first phylogenetic analysis of the whole genus of Trichodermausing sequence analysis of the ITS region of rDNA.
In this study, the method of isolation and identification of an unknown fungal from farmer field of Sultanpur district using 5.8 SrRNA gene sequence as reported in bacterial rRNA gene found by Srivastava, S. et al. (2008) to characterize the strain CA-03/9840as a member of the Trichoderma spp. The soil sample has received great attention from the public, due to its potential for biodiversity and biological conservation. The internal transcribed spacer (ITS) region of the rDNA is perhaps the most widely sequenced DNA region in fungi. It has typically been most useful for molecular systematic study at species level, and even within species Kindermann et al. (1998) attempted a first phylogeneticanalysis of the whole genus of Trichoderma using sequence analysis of the ITS1 region of rDNA.
Materials and methods
Isolation and identification of Trichoderma. Soil samples were collected from various experimental fields of Indian farmer field of Sultanpur district. Isolate of Trichoderma species was isolated and identified in potato dextrose agar (PDA) with low sugar medium reported by Hiney, M. et al. (1992). The identification of Trichoderma isolates were confirmed both by morphological and molecular characters (ITS) NCBI, Gene Bank Accession Number KU821782 and reconfirmed by Indian Type Culture Collection (ITCC), IARI, Pusa, New Delhi Accession Number allotted 9840 .
DNA isolation of Trichoderma. Pure culture of the target fungal was grown overnight in liquid PD Broth medium for the isolation of genomic DNA using a method described by Hiney, et al. (1998).
Molecular characterization. The total genomic DNA was extracted from isolate of Trichoderma asperellum CA-03/9840 based on Cetrimide Tetra decyl Trimethyl Ammonium Bromide (CTAB) mini extraction method of Crowhurst, et al. (1995) with minor modification.
Agarose gel electrophoresis. Ten microlitre of the reaction mixture was then analyzed by submarine gel electrophoresis using 1.0% agarose with ethidium bromide at 8 V/cm and the reaction product was visualized under Gel Doc/UV trans-illuminator.
Internal Transcribed Spacer region. The internal transcribed spacer (ITS) regions of the rDNA repeat from the 3’end of the 5.8S and the 5’end of the 5.8S gene were amplified using the two primers, ITS-1 and ITS-4 which were synthesized on the basis of conserved regions of the eukaryotic rRNA gene reported by Zhang, Z. (2000). The PCR-amplification reactions were performed in a 50 ml mixture containing 50 mMKCl, 20 mM Tris HCl (pH 8.4), 2.0 mM MgCl2, 200 mM of each of the four deoxy nucleotide triphosphates (dNTPs), 0.2µmM of each primer, 40 ng/ml of template and 2.5 U of Taq polymerase. The cycle parameters included an initial denaturation at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 1 min, primer annealing at 55°C for 2 min and primer extension at 72°C for 3 min and a final extension for 10 min at 72°C. Amplified products were separated on 1.2% agarose gel in TAE buffer, pre-stained with ethidium bromide (1mg/ml) and electrophoresis was carried out at 60 volts for 3 hours in TAE buffer. One Kb ladder (MBI, Fermentas) was used as a marker. The gel was observed in a trans-illuminator over ultra violet light. The desired bands were cut from the gel with minimum quantity of gel portion using QIAGEN gel extraction kit.
Purification of PCR product. The PCR product was purified by Qiagen gel extraction kit using the following protocol described below. The DNA fragment was excised from the agarose gel with a clean sharp scalpel. Then the gel slice was weighed in an eppendorf. We then added 3 volumes of buffer QG to 1 volume of gel (100 mg ~ 100 ?l). The mixture was then incubated at 50°C for 10 min. The gel was dissolved by vortexing the tube every 2 to 3 min during the incubation until the mixture color is uniformly yellow. We then added 1 gel volume of isopropanol to the sample and mixed. A QIA quick spin column is then placed in a 2 ml collection tube provided. The sample is applied to the QIAquick column followed by centrifugation for one minute so that DNA binds to the column. The flow-through is discarded and the QIA quick column is placed back in the collection tube. We then added 0.75 ml of buffer PE to QIA quick column and centrifuged for 1 min to wash. The flow through is again discarded and the QIA quick column centrifuged for an additional 1 min at 10,000 x g. The QIA quick column is now placed into a clean 1.5 ml eppendorf. We then added 50ml of buffer EB (10 mM Tris-Cl, pH 8.5) to the center of the QIA quick membrane and centrifuged the column for 1 min to elute DNA.
DNA sequencing of the 5.8SrDNA fragment. The 5.8SrDNA amplified PCR product (100 ng concentration) was used for the sequencing with the single 5.8SrDNA 20F Forward, ITS1 primer: 5′-TCCGTAGGTGAACCTGCGG-3’and 22R Reverse ITS4 primer: 5′-TCCTCCGCTTATTGATATGC-3’synthesized by DNA Sequencer by (Merck laboratory, Bangalore).
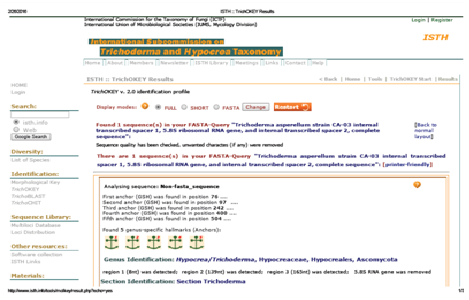
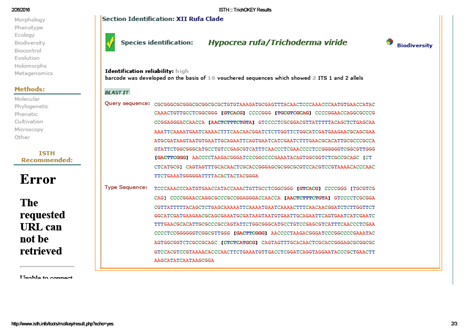
Fig. 2: TrichoKey analysis of Trichoderma asperellum strain CA-03
Sequence Analysis. A comparison of the 5.8SrRNA gene sequence of the test strain was done using BLAST against non-redundant nucleotide (nr/nt) database observed by Thompson, J.D. et al. (1994). A number of Trichodermasequences were selected on the basis of a similarity score of 90% with database sequences. Multiple sequence alignment of these selected homologous sequences and 5.8SrRNA gene sequence of test strain was performed using Clustal W reported by Saitou, N. et al. (1987). Subsequently, an evolutionary distance matrix was then generated from these nucleotide sequences in the dataset. A phylogenetic tree was then drawn using the Neighbour Joining method reported by Tamura, K. et al. (2007). Phylogenetic and molecular evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) version 4.0 reported by Altschul, S. F. et al. (1997). We again compared the 5.8SrRNA gene sequence of test strain with different set of sequence database such as small subunit ribosomal RNA (SSUrRNA) and large sub unitribosomal RNA (LSUrRNA) using Ribosomal RNA BLAST reported by Cole, J. R. (2009). 5.8SrRNA gene sequence of test strain is also compared against those sequences, in Ribosomal Database Project found by Wang, Q. et al. (2007) by using the RDP Classifier check Program. The annotated information for the sequence in the database to which 5.8SrRNA aligns is used for the fungal identification.
Results
A total of 573bp of the 5.8SrRNA gene was sequenced and used for the identification of isolated fungal strain. Subsequently, 5.8SrRNA gene sequence based phylogenetic tree showing therelationships between the test strain CA-03/9840 and selected representatives of the genus Trichoderma is given in Figure 1. It is evident from phylogenetic analysis of 5.8SrRNA gene that the isolate CA-03/9840 represents a genomic species in the genus Trichoderma. Comparison of teststrain against known sequences of SSUrRNA and LSUrRNA databases showed that the gene sequence of isolate CA-03/9840 has 90% sequence similarity (Score=546 bits, Expect=0.0) with 5.8SrRNA gene sequence of Trichoderma (Gene bank Acc. No.: KU821782). Thus, data shows that the isolate CA-03/9840 is a member of the genus Trichoderma. Similarity rank program classifier available at the Ribosomal Database Project classified the isolate CA-03/9840 as a novel genomic species of the genus Trichoderma with a confidence threshold of 90% (Figure 2). The 5.8SrRNA gene sequence of isolate CA-03/9840 was deposited in Gene Bank and allotted the accession number KU821782.
Discussion
Molecular analysis using Internal Transcribed Spacer region (ITS). Ribosomal RNA (rRNA) sequence analysis has been well-documented as a means of determining phylogenetic relationships in all of the major organismal domains. Variable sequences within the mature small subunit (SSU) and large-subunit (LSU) rRNA genes have been found to be appropriate for assessing sub-generic relationships in many eukaryotes. One of these variable regions, the D2 region of the LSU, has been used to determine phylogenetic has been used to determine phylogenetic relationships in a number of pathogenic fungal genera found by Logrieco, A. et. al. (1995). The ITS region of the rDNA operon, which is more variable than the D2 region, has proven useful in distinguishing relationships at the species level found by Kusaba, M. et al. (1995).
The genetic variability within 69 bio-control isolates of Trichoderma collected from different geographic locations and culture collections and their phylogenetic analysis were done with the help of the sequence data obtained from the inter Transcribed spacer 1(ITS1) region of Ribosomal DNA and a fragment of the translation elongation factor 1(tef1) and reported that more than 50% of the potential bio-control strains were grouped within Trichoderma section Pachybasium reported by Hermosa, M. et al. (2004).
Trichoderma isolates with different biocontrol capabilities and identification by molecular methods and further characterized into three main clades by internal transcribed spacer (ITS) sequence analyses. Consequently, a reliable phylogenetic tree was constructed containing isolates belonging to the T. harziunum clade reported by M. Maymon et al. (2004).
Molecular phylogenetic analyses of biological control strains of Trichoderma (Ascomycetes, Hypocreales) strains that have warted conidia are traditionally identified as T. viride, the type species of Trichoderma. However, two morphologically distinct types of conidial warts (I and II) have been found. Because each type corresponds to a unique mitochondrial DNA pattern, it has been questioned whether T. viride comprises more than one species. Combined molecular data (sequences of theinternal transcribed spacer 1 [ITS-1] and ITS-2 regions and of part of the 5.8SrRNA gene along with resultsof restriction fragment length polymorphism analysis of the endo-chitinase gene and PCR fingerprinting), morphology, physiology, and colony characteristics distinguish type I and type II as different species. Type I corresponds to “true” T. viride, the anamorphic of Hypocrearufa. Type II represents a new species, T. asperellum, which is, in terms of molecular characteristics, close to the neotype of T. hamatum.
Analysis of ITS1-5.8S-ITS2 region of the cDNA showed that approximate 600 bp and size variation was observed. Restriction analysis of this region showed that inter and intra -specific polymorphism found by Latha, J. et al. (2002).
Trichoderma has attained importance for substitute of chemical pesticides and hence an attempt was intended to corroborate the positive relatedness of molecular and morphological characters. A fungal strain of Trichoderma longibrachiatum 28CP/7444 was isolated from a soil sample collected from Barabanki district of Uttar Pradesh, India. The universal primers were used for amplification of the 28SrRNA gene fragment and strain characterized by using 28SrRNA gene sequence with the help of ITS marker. It is proposed that the identified strain Trichoderma longibrachiatum 28CP be assigned as the type strain of a species of the genus Trichoderma based on phylogenetic tree analysis together with the 28S rRNA gene sequence search in Ribosomal Database Project, small subunit rRNA and large subunit rRNA databases. The sequence was deposited in Gene Bank with the accession number JX978541. Thus, an integrated approach of morphological and molecular markers can be employed to identify a superior strain of Trichoderma for its commercial exploitation. Previously similar results were also reported by Shahid, M. et al. (2013, 2014) and they also concluded that most of the Trichoderma species are morphologically very similar and were considered as a single species for many years. Since new species were discovered, a consolidated taxonomical scheme was needed, proposed and defined nine morphological species aggregates. DNA methods brought additional valuable criteria to the taxonomy of Trichoderma which are being used today for studies that include identification and phylogenetic classification.
Acknowledgements
The authors are grateful to the financial support granted by the ICAR under the Niche Area of Excellence on “Exploration and Exploitation of Trichoderma as an antagonist against soil borne pathogens” running in Department of Plant Pathology,C.S.A University of Agriculture and Technology,Kanpur-208002, U.P., India.
References
- Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 1997; 25(17): 3389-402.
- Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., Kulam-Syed-Mohideen, A. S., McGarrell, D. M., Marsh, T., Garrity, G. M. and Tiedje, J. M. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis, Nucleic Acids Res., 2009; 37: D141-D145.
- Crowhurst, R. N., King, F. Y., Hawthorne, B. T., Sanderson, F. R. and Choi-Pheng, Y. RAPD characterization of Fusarium oxysporum associated with wilt of angsana (Pterocarpus indicus) in Singapore, Mycol. Res. 1995; 99: 14-18.
- Hermosa, M. R., Keck, E., Chamorro, I., Rubio, B., Sanz, L., Vizcaino, J. A., Gronodona, I. and Monte, E. Molecular characterization of bio-control agents, Bulletin-OILB-SROP, 2004; 27(8): 165-168.
- Hillis, D. M., Moritz, C., Porter, C. A. and Baker, R. J. Evidence for biased gene conversion in concerted evolution of ribosomal DNA, Science, 1991; 251: 308-310.
- Hiney, M., Dawson, M. T., Heery, D. M., Smith, P. R., Gannon, F. and Powell, R. DNA probe for Aeromonassalmonicida. Appl Environ Microbiol., 1992; 58(3):1039-42.
- Kindermann, J., Ayouti, Y. E., Samuels, G. J. and Kubicek, C. P. Phylogeny of the genus Trichoderma based on sequence analysis of the internal transcribed spacer region 1 of the rDNA cluster. Fungal GenBiol, 1998; 24: 298-309.
- Kubicek, C. P., Mach, R. L., Peterbauer, C. K. and Lorito, M. Trichoderma From genes to biocontrol. J Plant Pathol, 2000; 83: 11-23.
- Kulling-Gradinger, C. M., Szakacs, G. and Kubicek, C. P. Phylogeny and evolution of the genus Trichoderma: a multigene approach. Mycol Res, 2002; 106: 757-767.
- Kusaba, M., and Tsuge, T. Phylogeny of Alternaria fungi known to produce host-Specific toxins on the basis of variation in internal transcribed spacers of ribosomal DNA. Curr Genet, 1995; 28 : 491-498.
- Latha, J. and Mukherjee, P. K. Molecular characterization of ex-type strains of Trichoderma spp. from two Indian type cultures, collections. BARC Newsletter (founder’s day special issue) 2002; 145-149.
- Lee, C. and Hseu, T. Genetic relatedness of Trichoderma sect. Pachybasium species based on molecular approaches. Can J Microb, 2002; 48: 831-840.
- Logrieco, A., Peterson, S. W. and Bottalico, A. Phylogenetic relationships within Fusarium sambucinum Fuckelsensulato, determined from ribosomal RNA Sequences, Mycopathologia, 1995; 129: 153-158.
- Maymon, M., Minz, D., Barbul, O., Zveibil, A., Elad, Y., and Freeman, S. Identification of Trichoderma Biocontrol Isolates to Clades According to ap- PCR and ITS Sequencs Analysis Phytoparasitica, 2004; 32(4): 370-375.
- Nirenberg, H. I. Untersuchungenuber die morphologische und biologischedifferenzierung in der Fusarium Sektion Liseola. Mitt Biol Bundesanstalt fur Land-Forstw Berlin-Dallem, 1976; 169: 1-117.
- Ospina-Giraldo, M. D., Royse, D. J., Thon, M. R., Chen, X. and Romaine, C. P. Phylogenetic relationships of Trichoderma harzianum causing mushroom green mold in Europe and North America to other species of Trichoderma from world-wide sources, Mycologia, 1998; 90: 76-81.
- Sacchi, C. T., Whitney, A. M., Reeves, M. W., Mayer, L. W., Popovic, T. Sequence Diversity of Neisseriameningdidis 16S rRNA Genes and Use of 16S rRNA Gene Sequencing as a Molecular Subtyping Tool. J Clin Microbiol, 2002; 40(12): 4520-4527.
- Saitou, N. and Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol, 1987; 4(4): 406-25.
- Shahid, M., Singh, A., Srivastava, M., Rastogi, S. and Pathak, N. Sequencing of 28S rRNA gene for identification of Trichoderma longibrachiatum 28CP/7444 species in soil sample. International Journal of Biotechnology for Wellness Industries, 2013; 2: 84-90.
- Shahid, M., Srivastava, M., Sharma, A., Singh, A., Pandey, S., Kumar, V., Pathak, N. and Rastogi, S. Sequencing of 28s rRNA gene for Identification of Trichoderma longibrachiatum 21PP species in soil sample. African Journal of Microbiology Research, 2013; 7: 4902-4906.
- Shahid, M., Srivastava, M., Kumar, V., Singh, A., Sharma A., Pandey, S., Rastogi, S. and Srivastava, A. K. Phylogenetic Diversity Analysis of Trichoderma species Based on ITS Marker. African Journal of Biotechnology, 2014; 13(3): 449-455.
- Srivastava, S., Singh, V., Kumar, V., Verma, P. C., Srivastava, R., Basu, V., Gupta, V., and Rawat, A. K. Identification of regulatory elements in 16S rRNA gene of Acinetobacter species isolated from water sample. Bioinformation, 2008; 3(4):173-176.
- Tamura, K., Dudley, J., Nei, M. and Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol Evol, 2007; 24(8):1596-99.
- Thompson, J. D., Higgins, D. G. and Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res, 1994; 22(22): 73-80.
- Wang, Q., Garrity, G. M., Tiedje, J. M. and Cole, J. R. Na7ve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol, 2007; 73(16): 61-67.
- White, T. J., Bruns, T., Lee, S. and Taylor, J. Amplification and direct sequencing of fungal ribosomal DNA for phylogenetics. In: Innes MA, Gelfand DH, Sninsky JJ, White TJ (Eds.), PCR protocols: a guide to methods and applications. Academic Press, Inc, San Diego, Calif 315-322 pp 1990.
- Woese, C. R., Fox, G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA, 1997; 74: 5088-5090.
- Woese, C. R. Bacterial evolution. Microbiol Rev, 1987; 51: 221-271.
- Zhang, Z., Schwartz, S., Wagner, L. and Miller, W. A greedy algorithm for aligning DNA sequences. J Comput Biol, 2000; 7(1-2):203-14.

