ISSN: 0973-7510
E-ISSN: 2581-690X
Three keratinolytic bacterial isolates were characterized partially for their keratinase activity. Bacterial isolates were grown in feather meal agar. Ammonium sulfate precipitation followed by dialysis was performed to know the bacterial isolate keratinase activity in differet pH and temperature. Identification of the bacteria was done by using their 16S rRNA gene sequences. The result showed that bacterial growth was coinciding with keratinase activity. Precipitation with ammonium sulfate showed that keratinae activity of isolate A4 was optimum at 20% of ammonium sulphate, while B4 and B6 were more active at 70%. Keratinase activity increased after dialysis. Keratinase of A4 showed to have optimum activity at temperature of 45oC and pH=8, B4 was optimum at temperature of 35oC and pH=7, while B6 was optimum at temperature of 40oC and pH=7, respectively. Identification of the bacterial isolates using 16S rRNA gen showed that A4, B4, and B6 were closed to Leclercia adecarboxylata strain M-X17B, Azotobacter chroococcum strain ABA-1, and Stenotrophomonas maltophilia strain BIW by 97%, 99%, and 98%, respectively. Two bacteria L. adecarboxylata and A. chroococcum were firstly reported to produce keratinase.
Keratinase, Chikhen feather, Goat fur, Azotobacter chroococcum, Leclercia adecarboxylata, Stenotrophomonas maltophilia.
Poultry processing plants are producing millions of tons of feathers, which mainly consisted of keratin (Brandelli, 2008; Agrahari & Wadhwa, 2010; Mazotto et al., 2010; El-Ayouty et al., 2012). Keratin filament structures are stabilized by their high degree of cross-linking of disulfide bonds, hydrophobic interactions, and hydrogen (Brandelli, 2008; Xu et al., 2009), which make them unsoluble and resistant to most of the known proteases like trypsin, pepsin, papain and results in polluting the environment (Brandelli, 2008; Xu et al., 2009; Kansoh et al., 2009; Duarte et al., 2011; Sivakumar et al., 2012). However, keratin does not accumulate in nature (Rahayu et al., 2010).
Keratin sources such as feather, horn, nails and hair are abundantly available in nature as wastes. High protein content of keratin waste can be used as a good source of protein and amino acids by systemic recycling, which may provide a cheap and alternative protein feed stuff (Jeong et al., 2010; Duarte et al., 2011; Mazotto et al., 2011; Tiwary & Gupta, 2012). Feathers are currently used to produce feather meal using physical and chemical treatments, destroying certain amino acids and resulting in a low nutritional value product (Jeong et al., 2010; Duarte et al., 2011; Mazotto et al., 2011; Tiwary & Gupta, 2012).
Certain microorganisms produce keratinases to degrade keratins and may be used to enhance the digestibility of feather keratin (Riffel & Brandelli, 2008). The nutritional upgrading of feather meal through microbial or enzymatic treatment has been described (Xu et al., 2009; Agrahari & Wadhwa, 2010; El-Ayouty et al., 2012). Bacterial genera such as Burkholderia, Chryseobacterium, Pseudomonas, Microbacterium sp. (Riffel & Brandelli, 2008), Streptomyces (Brandelli, 2008), and Bacillus (Mazotto et al., 2011; Brandelli, 2008; Lucas et al., 2003) have been reported to synthesize keratinase. The bacteria were grown on substrats such as feather meal, raw feathers, chicken nails, hair, and wool as sole carbon and nitrogen sources for the bacterial growth (Riffel & Brandelli, 2008).
So far, little attention has been given in study of utilization and degradation of feather waste by keratinolytic microorganisms in Indonesia. Rahayu et al. (2010) reported a study on keratinase of two Indonesian bacterial isolates. In this study, partial characterization of keratinolytic activity of three bacterial isolates from a feather decomposting waste was conducted. Their ability to degrade chikhen feather and goat hair has also been studied.
Cell growth and keratinase activity
Bacterial culture was spreaded on feather meal agar (0,5 g/l NaCl; 0,7 g/l K2HPO4; 1,4 g/l KH2PO4; 0,1 g/l MgSO4 added with 10 g/l feather meal, and 15 g/l agar) and incubated at ambient temperature (Cai et al., 2008). Cell growth was measured everyday as colony forming unit/ml.
To measured keratinase activity, 500 µl crude enzyme was added with of phosphate buffer saline (PBS) of pH 7, and mixed with 500 µl of 0.5% keratin solution. Enzymatic reaction was stopped by adding 1 ml of 10% tricloroacetic acid, and spinned at 10.000 rpm for 30 minutes. Free amino acid in supernatant was measured using spectrophotometer at ë=280 nm. One unit of enzyme activity is defined as the increasing of absorbance of 0.01.
Ammonium sulphate precipitation and dialysis
Broth culture of bacterial isolates was centrifugated at 10.000 g for 10 minutes. Supernatant was precipitated using 20 to 70% of ammonium sulphate for 2 hours, and was spinned at 10.000 rpm for 10 minutes at 4oC. Precipitate and supernatant was subjected to keratinase activity assay. Precipate was solubilized with PBS of pH 7 prior enzyme activity assay. Precipitate showed higher activity was choosen for dialysis. Dialysis was conducted using PBS for 6 hours by changing the buffer twice. Keratinase activity of dialysis was measured as previously described.
Characterization of keratinase in different pH and temperature
Dialysed enzyme was subjected to be assayed in different temperature and pH. To characterize enzyme temperature, enzyme and substrate mixture was incubated at 31°C of optimum pH for 15 minutes. Effect of pH on keratinase activity was measured at varying pH of 4.0, 5.0, with Na-acetate buffer, 6.0, 7.0 with K-phosphate buffer, and 8.0, 9.0 with Tris-HCl buffer. The reaction was conducted at 31°C for 15 minutes. Keratinase activity was measured as previously described.
Identification of keratinolytic bacterial isolates based on its 16S rRNA genes
Keratinolitik bacterial DNA isolation WAS done by freeze and thaw method. One loop of bacterial culture1 of 24 hours put into microtube of 100 mL aquabidest. Suspension was frozen at -10oC and thawed with 90°C for 10 minutes for 5 times. Suspension was spinned and supernatan was taken out. The supernatan was used for 16S rRNA gene amplication using Ready To Go PCR Beads (Pharmacia-Biothec) with primer 63f (5’-CAGGCCTAACACATGCAAGTC-3’) dan 1387r (5’- GGCGGWGTGTACAAGGC-3’) (Marchesi et al., 1998).
Total volume of the PCR reaction (25 ml) consisted of 1.5 U Taq DNA polymerase, 10mM Tris-HCl (pH 9 at room temperature), 50 mM KCl, 1.5 mM MgCl2, 200 mM of each dNTPs and stabilizer including BSA. Reaction was conducted for pradenaturation at 94oC for 2 minutes, denaturation at 92oC for 30 seconds, annealing at 55oC for 30 seconds, elongation at 72oC fro 1 minute, and post-PCR 72oC for 5 minutes in a thermocycler (Verity® 96-Well Thermal Cycler 437586, Applied Biosystems, Singapore). PCR was done for 30 reaction. PCR result was visualised with mini gel electrophoresis.
Construction of phylogenetic tree
Amplified 16S rRNA gene was purified and commercially sequenced. DNA sequence was compared to GenBank database of The National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) using Basic Local Alignment Search Tool (BLAST). Cluster analysis was conducted using MEGA7.
Cell growth and keratinase activity
Keratinase production profiles of each bacterial isolates were not similar. B4 showed to have higher cell number and keratinase activity at 5 days of incubation, while the other two A4 and B6 were higher at 4 days of incubation. However, it seemed that keratinase activity was in line with cell growth (Figure 1.). Maximum activity of keratinase of three isolates was shown at the end of exponential growth phase and at stasioner phase. Werlang & Brandelli (2005) reported that maximum keratinase activity of Bacillus sp. kr 16 incubated at 30ºC occurred at the end of exponential growth phase, as also showed by Zhang et al. (2009) of Bacillus sp. 50-3 incubated at 37ºC. Cell growth and keratinase production could be increased by using C-source such as starch (Gioppo et al., 2009).
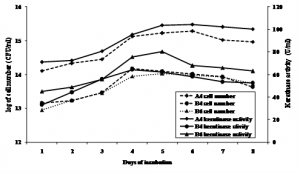
Fig. 1. Keratinase activity during bacterial cell growth in feather meal broth at 31°C and initial pH 6.5
Ammonium sulphate precipitation
Keratinase activity of precipitate and supernatant were measured after ammonium sulphate precipitation. It was observed that keratinase activity of fraction of 40 to 60% of ammonium sulphate were relative similar between pellet and supernatant of all isolates. Keratinase activity of pellet of B4 was higher at 70%. Interestingly, keratinase of the isolotes showed to have two peaks. Keratinase of A4 and B6 was optimum at 20 and 70%, while that of B4 was optimum at 30% and 70%, respectively (Figure 2.). This result indicated that the isolates might produce two different proteins. Rahayu et al., (2010) reported that Bacillus sp. MTS showed to produce 6 different proteins of 17, 25, 32, 53, 96 and 122 kDa showing keratinolityc activity.
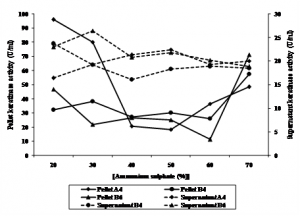 Fig. 2. Keratinase activity of pellet and supernatant of bacterial culture precipitated using ammonium sulphate
Fig. 2. Keratinase activity of pellet and supernatant of bacterial culture precipitated using ammonium sulphateFraction of 20% of A4 protein, and of 70% of B4 and B6 were subjected to dialysis. Specific activity of keratinase increased after ammonium sulphate precipitation and dialysis, while total protein decreased, as shown in Table 1. Bacillus pseudofirmus keratinase activity precipited with ammonium sulphate increased its keratinase activity to 55.68 U/ml (Kojima et al., 2006). Keratinase activity of isolat L1 precipited with ammonium sulphate followed with dialysis increased 3 fold with yield of 42% (Cao et al., 2009).
Table (1):
Purification of keratinase produced by the keratinolytic bacterial isolalates.
| Isolates | Purification step | Total protein (mg/ml) | Enzyme activity (U/ml) | Spesific activity (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| A4 | Supernatant | 55.41 | 108.70 | 1.96 | 100 | 1 |
| Ammonium sulphate (20%) | 38.88 | 95.90 | 2.47 | 88.22 | 1.26 | |
| Dialysis | 21.64 | 99.70 | 4.61 | 91.72 | 2.35 | |
| B4 | Supernatant | 52.18 | 65.80 | 1.26 | 100 | 1 |
| Ammonium sulphate (70%) | 42.81 | 57.50 | 1.34 | 87.39 | 1.07 | |
| Dialysis | 20.20 | 63.20 | 3.13 | 96.05 | 2.48 | |
| B6 | Supernatant | 66.79 | 86.80 | 1.30 | 100 | 1 |
| Ammonium sulphate (70%) | 49.22 | 71.20 | 1.45 | 82.03 | 1.11 | |
| Dialysis | 20.54 | 83.90 | 4.08 | 96.66 | 3.14 |
Effect of pH on keratinase activity
Optimum pH of A4, B4, and B6 keratinase activity varied. B4 and B6 showed to have optimum activity at pH 7, while A4 was optimum at pH 8. Keratinase activity declined at pH above or below 7 and 8 (Figure 3.). Keratinase Bacillus subtilis MTCC (9102) (Kumar et al., 2010), Stenotrophomonas maltophilia DHHJ (Cao et al., 2009), and B. subtilis (Cai & Zheng, 2009), was optimum at pH 7, 7.8, and 8.5, respectively. Werlang & Brandelli (2005) reported that their bacterial isolate keratinase were active at pH 8 to 11, while Bacillus sp. 50-3 (Zhang et al., 2009) and B. halodurans PPKS-2 (Prakash et al., 2010) showed to have optimum pH of 10 and 11, respectively.
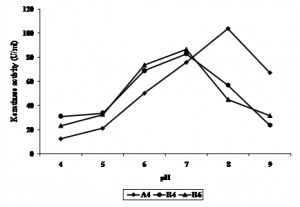 Fig. 3. Effect of pH on keratinase activity of bacterial isolates
Fig. 3. Effect of pH on keratinase activity of bacterial isolatesEffect of temperature on keratinase activity
Keratinase activity of all isolates showed to have different activity at different temperature (Figure 4.). The optimum temperature for A4 was at 45°C, B4 was at 35°C, while B6 was at 40°C. Keratinase of Bacillus kr 16 (Werlang & Brandelli, 2005) and B. halodurans PPKS-2 (Prakash et al., 2010) of chikhen feather waste were optimum at 45-65oC and 60-70oC, respectively. Zhang et al., (2009) reported keratinase of Bacillus sp 50-3 isolated from animal feces of Beijing Zoo was optimum at 60oC. Keratinase of Paracoccus sp. isolated from poultry soil was optimum at 50oC (Lee et al., 2004). Stenotrophomonas maltophilia DHHJ (Cao et al., 2009) and B. subtilis MTCC (9102) (Balaji et al., 2008) showed their optimum keratinase activity at 40oC, while B. subtilis (Cai & Zheng, 2009) and isolate kr 6 (Riffel et al., 2003) were more active at 55oC.
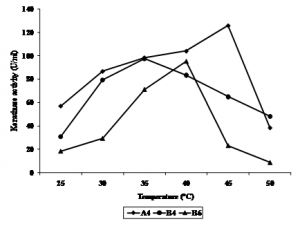 Fig. 4. Effect of temperature (°C) on keratinase activity of bacterial isolates
Fig. 4. Effect of temperature (°C) on keratinase activity of bacterial isolates
Identification of keratinolytic bacterial isolate based on its 16S rRNA gene
Isolates A4, B4, and B6 were identified by using their 16S rRNA gene sequences. A4, B4, and B6 was closely related to Leclercia adecarboxylata strain M-X17B, Azotobacter chroococcum strain ABA-1, and Stenotrophomonas maltophilia strain BIW by 97%, 99%, and 98%, respectively (Figure 5.). Leclercia adecarboxylata and Azotobacter chroococcum has not been reported previously as keratinolytic bacteria. So far, this is the first report that these two bacteria produced keratinase. Leclercia adecarboxylata was previoulsy known as opportunistic pathogenic bacteria (Keren et al., 2014), and was utilised as biocontrol agents of Leptinotarsa decemlineata (Muratoglu et al., 2009). It was also reported to degrade polycyclic aromatic hydrocarbons of oil spills contaminated soil (Sarma et al., 2004). Azotobacter chroococcum was reported as nirogen fixing bacteria (Mahato et al., 2009; Damir et al., 2011) and in producing polyester polyhydroxialkanoic (Lopez et al., 1996).
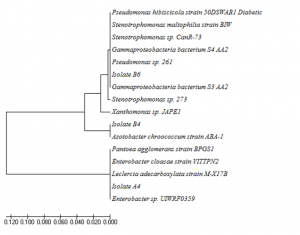
Fig. 5. Phylogenic tree of isolat A4, B4, and B6 based on their 16S rRNA genes
One of the isolate B6 was identified as Stenotrophomonas maltophilia. This bacteria was isolated from chikhen feather composting soil in China. Growth and degradation activity of S. maltophilia DHHJ were optimum at at 40ºC in pH of 7,5-8 (Cao et al., 2009).
Keratin is a fibrous and insoluble structural protein found in feathers, hair, nails, horns and other epidermal tissue. Extensively cross-linked by disulfide, hydrogen and hydrophobic bonds makes it very resistant to digest by common proteases like pepsin, papain, dan tripsin (Werlang & Brandelli, 2005). Keratin can be degraded by breaking or break the disulfide bonds in keratin. This can be done with high heat (hydrothermal), chemical treatment and biological treatment. Hydrothermal treatment is done by setting a high temperature and pressure, as well as chemical treatments carried out by the addition of acid (HCl) and base (NaOH) at a high concentration (Cai & Zheng, 2009).
Many bacteria have been reported to produce keratinase. Keratinase production profile of each microorganism can vary. It depends on the gene encoding the keratinase, the composition of culture medium as a substrate, concentration of carbon and nitrogen, temperature, pH, and aeration (Brandelli et al., 2010). Optimum temperature of bacterial enzyme activity is usually correlated with temperature in where the bacteria live. All of our isolates were isolated from chikhen feather waste which was progressively decomposed. The bacteria live in should be appropriate ones, having optimum enzyme acitivity of higher temperature, as also shown, for examples in Werlang & Brandelli (2005), Prakash et al. (2010), Zhang et al., (2009), Lee et al., (2004), and Cao et al. (2009) who isolated the bacteria from decomposing chikhen feather, animal feces, and poultry soil. Our keratinase might be induced by keratin substrat such as feather and fur, since the isolates were able to grow and produced keratinase in medium in which feather and fur served as a sole carbon and nitrogen source, as also shown by Suntornsuk et al. (2005).
Most keratinases are alkaline or netral protease with optimum pH of 7.5 to 9. However, some were extremely acidic or alkaline (Brandelli et al., 2010). Kumar et al., (2010), Prakash et al., (2010), Cao et al., (2009), Cai & Zheng (2009), Zhang et al., (2009), and Werlang & Brandelli (2005) showed that their bacterial keratinase activities were optimum at pH between 7 to 11. Our isolates showed to be more active in slightly alkaline or netral with optimum pH of 7 to 8, accordingly.
The keratinase of A4 and B6 reached maximum production in 5 days, while B4 reached it in 4 days, in which maximum growth was observed. This was similar to that of Suntornsuk et al. (2005) in which the maximum production of keratinase reached a maximum in the late logarithmic growth phase. Based on the maximum growth that were coinciding with maximum biomass and maximum specific production rates observed at the exponential growth phase, our keratinases were produced as a primary metabolite (Brandelli & Riffel, 2005). After reach maximum cell growth and enzyme activity, a loss of enzyme activity was observed probably because of enzymatic autolysis and end product inhibition (Suntornsuk et al., 2005).
The isolates showed only partial digestion of both chikhen feather and goat fur, with more chikhen feather degraded. Kansoh et al. (2009) showed that feather was more easy to be degraded compared tho that of hair, nail, and wool. This result indicated that this enzyme could not completely cleave the disulphide bonds. Pre-treatment of these substrates such as chicken feather, nail and human hair by physical method or reducing agents, or detergents, or activation of the enzyme by adding metal salts are required for the improvement of their degradation (Suntornsuk et al., 2005).
ACKNOWLEDGMENTS
We would like to thank to DRPM, Ministry of Research, Technology, and Higher Education, Republic of Indonesia to fully supporting this research.
- Agrahari S, Wadhwa N. Degradation of chicken feather a poultry waste product by keratinolytic bacteria isolated from dumping site at Ghazipur poultry processing plant. Int J Poultry Scie. 2010; 9: 482-489.
- Balaji S, Kumar MS, Karthikeyan R, Kumar R, Kirubanandan S, Sridhar R, Sehgal PK. Purification and characterization of an extracellular keratinase from a hornmeal-degrading Bacillus subtilis MTCC (9102). World J Microbiol Biotechnol.2008; 24: 2741–2745.
- Brandelli A. Bacterial keratinases: Useful enzymes for bioprocessing agroindustrial wastes and beyond. Food Bioprocess Technol., 2008; 1: 105-116.
- Brandelli A, Daroit DJ, Riffel A. Biochemical features of microbial keratinases and their production and applications. Appl Microbiol Biotechnol. 2010; 85: 1735-1750.
- Cai C, Zheng X. Medium optimization for keratinase production in hair substrate by a new Bacillus subtilis KD-N2 using response surface methodology. J Ind Microbiol Biotechnol. 2009; 36: 875–883.
- Cao Z, Zhang Q, Wei D, Chen L, Wang J, Zhang X, Zhou M. Characterization of a novel Stenotrophomonas isolate with high keratinase activity and purification of the enzyme. J Ind Microbiol Biotechnol. 2009; 36: 181-188.
- Damir O, Mladen P, Bozidar S, Srdan N. Cultivation of the bacterium Azotobacter chroococcum for preparation of biofertilizers. Afri J Biotechnol. 2011; 10(16): 3104-3111.
- Duarte TR, Oliveira SS, Macrae A, Cedrola SML, Mazotto AM, Souza EP, Melo ACN, Vermelho AB. Increased expression of keratinase and other peptidases by Candida parapsilosis mutants. Braz J Med Biol Res. 2011; 44: 212-216.
- El-Ayouty YM, El-Said A, Salama AM. Purification and characterization of a keratinase from the feather-degrading cultures of Aspergillus flavipes. Afr J Biotechnol. 2012; 11: 2313-2319.
- Gioppo NM, Moreira FG, Costa AM, Alexandrino AM, De Souza CG, Peralta RM. Influence of the carbon and nitrogen sources on keratinase production by Myrothecium verrucaria in submerged and solid state cultures. J Ind Microbiol Biotechnol. 2009; 36: 705-711.
- Jeong E-J, Rhee M-S, Kim G-P, Lim K-H, Yi D-H, Bang B-H. Purification and characterization of a keratinase from a feather-degrading bacterium, Bacillus sp. SH-517J. Korean Soc Appl Biol Chem. 2010; 53: 43-49.
- Kansoh AL, Hossiny EN, El-Hameed EKA. Keratinase production from feathers wastes using some local Streptomyces isolates. Australian J Basics Appl Scie. 2009; 3: 561-571.
- Keren Y, Keshet D, Eidelman M, Geffen Y, Raz-Pasteur A, Hussein K. Is Leclercia adecarboxylata a New and Unfamiliar Marine Pathogen? J Clin Microbiol., 2014; 52(5): 1775-1776.
- Kojima M, Kanai M, Tominaga M, Kitazume S, Inoue A, Horikoshi K. Isolation and characterization of a feather-degrading enzyme from Bacillus pseudofirmus FA30-01. Extremophiles. 2006; 10: 229–235.
- Kumar R, Balaji S, Uma TS, Mandal AB, Sehgal PK. Optimization of influential parameters for extracellular keratinase production by Bacillus subtilis (MTCC9102) in solid state fermentation using horn meal-a biowaste management. Appl Biochem Biotechnol. 2010; 160: 30-39.
- Lee YJ, Kim JH, Kim HK, Lee JS. Production and characterization of keratinase from Paracoccus sp. WJ-98. Biotechnol Bioproc Engin. 2004; 9: 17-22.
- Lopez JG, Pozo MV, Toledo M, Rodelas B, Salmeron V, Production of polyhydroxyalkanoates by Azotobacter chroococcum H23 in wastewater from olive oil mills (Alpechin). Int Biodeterio Biodeg. . 1996; 96: 271-276.
- Lucas FS, Broennimann O, Febbraro I, Heeb P. High diversity among feather-degrading bacteria from a dry meadow soil. Microb Ecol. 2003; 45: 282-290.
- Mahato P, Badoni A, Chauhan JS. Effect of Azotobacter and nitrogen on seed germination and early seedling growth in tomato. Researcher, 2009; 1(4): 62-66.
- Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Dymok D, Wade WG. Design and evaluation of useful bacterium-spesific PCR primers that amplify genes coding for bacteria 16s rRNA. Appl Environ Microbiol. 1998; 64(2): 795-799.
- Mazotto AM, de Melo ACN, Macrae A, Rosado AS, Peixoto R, Cedrola SML, Couri S, Zingali RB, Villa ALV, Rabinovitch L, Chaves JQ, Vermelho AB. 2010. Biodegradation of feather waste by extracellular keratinases and gelatinases from Bacillus spp. World J. Microbiol. Biotechnol. DOI 10.1007/s11274-010-0586-1
- Muratoglu H, Kati H, Demirbag Z, Sezen K. High insecticidal activity of Leclercia adecarboxylata isolated from Leptinotarsa decemlineata (Col.: Chrysomelidae). Afri J Biotechnol. 2009; 8(24): 7111-7115.
- Prakash P, Senigala K, Jayalakshmi, Sreeramulu K. Purification and characterization of extreme alkaline, thermostable keratinase and keratin disulfide reductase produced by Bacillus halodurans PPKS-2. Appl Microbiol Biotechnol. 2010; 87: 625-633.
- Rahayu S, Syah D, Suhartono MT. A preliminary study on keratinase from two Indonesian isolates. Animal Produc. 2010; 12: 60-68.
- Riffel A, Brandelli A. Keratinolytic bacteria isolated from feather waste. Brazilian J Microbiol. 2008; 37: 395-399.
- Riffel A, Lucas F, Heeb P, Brandelli A. Characterization of a new keratinolytic bacterium that completely degrades native feather keratin. Arch Microbiol. 2003; 179: 258-265.
- Sarma PM, Bhattacharya D, Krishnan S, Lal B. Degradation of polycyclic aromatic hydrocarbons by a newly discovered enteric bacterium, Leclercia adecarboxylata. Appl Environ Microbiol. 2004; 70(5): 3163-3166.
- Sivakumar T, Shankar T, Ramasubramanian V. Purification properties of Bacillus thuringiensis TS2 keratinase enzyme. American-Eurasian J Agric Environ Scie. 2012; 12: 1553-1557.
- Suntornsuk W, Tongjun J, Onnim P, Oyama H, Ratanakanokchai K, Kusamran T, Oda K. Purification and characterisation of keratinase from a thermotolerant feather-degrading bacterium. World J Microbiol Biotechnol. 2005; 21: 1111-1117.
- Tiwary E, Gupta R. Rapid conversion of chicken feather to feather meal using dimeric keratinase from Bacillus licheniformis ER-15. J Bioproces Biotechniq. 2012; 2(4): 5 pp. http://dx.doi.org/10.4172/2155-9821.1000123.
- Werlang PO, Brandelli A. Characterization of a novel feather degrading Bacillus sp. strain. Appl Biochem Biotechnol. 2005; 120: 71-79.
- Xu B, Zhong Q, Tang X, Yang Y, Huang Z. Isolation and characterization of a new keratinolytic bacterium that exhibits significant feather-degrading capability. Afri J Biotechnol. 2009; 8: 4590-4596.
- Zhang B, Jiang D, Zhou W, Hao H, Niu T. Isolation and characterization of a new Bacillus sp. 50-3 with highly alkaline keratinase activity from Calotes versicolor faeces. World J Microbiol Biotechnol. 2009; 25: 583-590.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


