ISSN: 0973-7510
E-ISSN: 2581-690X
This study investigates the plant growth-promoting (PGP) characteristics and biodegradation capacity of organophosphate-tolerant soil bacteria isolated from vegetable farms in Benguet, Philippines. Three soil bacterial strains identified based on 16S rRNA sequences as Pseudomonas chlororaphis (M4C4-5), Pantoea allii (M1C5-1), and Mammaliicoccus sciuri (M1C4-15) tolerated up to 2,000 mg L-1 chlorpyrifos (CP) and malathion (MT) and exhibited PGP characteristics including nitrogen (N2) fixation, ammonia (NH3) production, phosphate (PO43-), and potassium (K) solubilizations, and productions of indole-3-acetic acid (IAA), siderophores, and gibberellin A3 (GA3). Growth analysis using minimal media with CP as the sole carbon source suggests a maximum population (10.11 log10 CFU) on day 5, achieved by P. chlororaphis (M4C4-5). The isolates showed degradation of the amended CP in the mineral salt medium. Remarkably, P. chlororaphis (M4C4-5) showed efficient degradation by 81.76%. Overall, this study provided new insights and application potential of novel soil bacteria in the bioremediation of OP pesticides while promoting plant growth and development by producing phytohormones and enzymes.
Plant Growth-promoting Bacteria (PGPB), Biodegradation, Organophosphate Pesticides, Soil Microbial Diversity, Bioremediation, Chlorpyrifos
To cope with high food demands, the agricultural sector has doubled pesticide utilization and applications for pest control and management in 2020.1 In that year alone, approximately 2.7 metric tons (Mt) of pesticide active ingredients were used across the globe from more than 1,000 known pesticides.2,3 However, this massive and extensive pesticide dependency has resulted in its diffusion to the environment and the food chain.4
Researchers have documented and reported the pesticide utilization and contamination of agricultural soil, irrigation water, and crop produce in Benguet, known as the vegetable basket of the Philippines. Researchers detected carbamates, a toxic pesticide that targets cholinesterase enzymes (ChEs) in agricultural soil and river water samples collected.5 Similarly, tests found cyhalothrin (0.07 mg L-1) and chlorpyrifos (0.26 mg L-1) in agricultural soil and strawberries, both exceeding the maximum residue level (MRL) of 0.01 0.26 mg L-1.6 Researchers also identified 18 organophosphate pesticide (OP) residues in vegetable samples (cabbage, potato, and sweet pea) collected in Mt. Province and Benguet, all of which exceeded the MRL.7 Reports further highlighted that endosulfan sulfate, chlorpyrifos, cypermethrin, malathion, profenofos, chlorothalonil, cyhalothrin, fenvalerate, and deltamethrin were the most prevalent pesticides in all municipalities in Benguet, contaminating agricultural soils in Kapangan, and Buguias, Benguet.6,8
To address the pesticide residue problem in agricultural soils, indigenous microbial populations that have evolved a capacity for degrading these pesticides are being sought as more efficient and less expensive agents for the bioremediation of contaminated areas.9 Various researchers have studied and documented microorganisms such as Bacillus pumilus, Achromobacter xylosoxidans, Pseudomonas nitroreducens, Pseudomonas aeruginosa, Streptomyces sp., Paracoccus sp., Klebsiella sp., Alcaligenes sp., and Ochrobactrum sp. because of their potential in the microbial degradation of pesticides in the soil.10-12 In addition to degrading pesticides, these microbes can also contribute to crop development and growth. For instance, Arthrobacter oxydans and Bacillus flexus were identified as CP-tolerant soil bacteria isolated from potatoes that simultaneously increased the growth and phosphorus content of the potatoes.13 Additionally, Acinetobacter calcoaceticus was found to exhibit PGP characteristics like solubilization of phosphates and production of IAA and siderophores while breaking down and utilizing CP as its carbon source.14 The ability to leverage plant growth under conditions of pesticide and other abiotic stresses is due to their ability to produce plant hormones, scavenge reactive oxygen species, and produce exopolysaccharide substances.15,16 This is important in creating a favorable environment for crops to grow while reducing pesticide residues in soil and plant tissues.17
Before the in situ application of bioremediation strategies for agricultural lands, it is important to isolate, screen, and characterize indigenous soil bacterial strains that can degrade pesticides. This research focuses on uncovering these potential pesticide biodegraders from vegetable areas in Benguet, Philippines, and screening for their plant growth-promoting features.
Commercial-grade OP pesticides SIGA 300 EC (chlorpyrifos) and AGWAY Malathion were procured from local agricultural shops in Benguet. All other chemicals and culture media were purchased from Ajax Chemicals, Sigma-Aldrich, Merck, and HiMedia Laboratories.
Isolation, screening, and characterization of CP and MT-tolerant soil bacteria
Composite soil samples (1 kg each) were collected from fifteen (15) vegetable farms in four (4) municipalities of Benguet: Mankayan, Buguias, Kapangan, and La Trinidad. The collected soil samples were labeled for easy tracking, placed in an ice box, and immediately transported to the Research Laboratory of the Biology Department at Benguet State University for processing.
To isolate CP and MT-tolerant soil bacteria, 10 g of a fresh soil sample was homogenized, and 100 µl of the 10-4 and 10-5 dilutions were plated on minimal agar medium (MAM) supplemented with 100 mg L-1 CP and MT as the starting concentration of OP. The plates were incubated at 30 ± 2 °C for 48 hours.10,12 Pure cultures were kept on NA plates for cultural and morphological characterization.
Tolerance test of soil bacteria to increasing concentrations of CP and MT
Unique isolates were subjected to tolerance assessment using MAM plates with increasing concentrations of CP and MT: 200 mg L-1, 300 mg L-1, 400 mg L-1, 500 mg L-1, 1000 mg L-1, 1500 mg L-1, and 2000 mg L-1. The isolates were spot-inoculated and incubated at 30 ± 2 °C for 48 hours.10 After the incubation period, only the isolates that exhibited growth and tolerance to 2000 mg L-1 of either CP or MT were characterized for plant growth-promoting potentials.
Screening for Plant Growth-Promoting (PGP) characteristics
Nitrogen (N2) fixation and ammonia (NH3) production
Isolates were spot-inoculated on Jensen’s agar plates (nitrogen-free medium) and incubated at 30 ± 2 °C for 7 days. Growth on the media indicates capacity for nitrogen fixation.18 To validate the result of the nitrogen-fixing characteristics of the isolate, the production of ammonia (NH3) was tested. Ten ml of peptone water was inoculated with the nitrogen-fixing isolates in triplicate and incubated at 35 ± 2 °C for 96 hours. After the incubation period, 2 ml of culture broth of the bacteria was centrifuged at 14,500 x g for 5 minutes. One ml of the isolate’s supernatant was mixed with 500 ml of Nessler’s reagent and incubated at room temperature for 10 minutes. Absorbance was measured using the UV-Vis spectrophotometer (PG Instruments©, United Kingdom) at 450 nm. Uninoculated peptone water was used as a reference/blank solution. All readings were then averaged. The concentration of the ammonia was calculated based on the calibration curve of the ammonium sulfate (NH4)2SO4 solution.19
Phosphate (PO43-) and potassium (K) solubilization
Bacterial isolates were spotted separately on Pikovskaya’s (PO43-) and Aleksandrov’s (K) agar plates. Uninoculated agar plates served as the negative control. Pikovskaya’s agar plates were incubated at 35 ± 2 °C for 24 hours, while Aleksandrov’s agar plates were incubated at 30 ± 2 °C for 7 days.17 After the incubation periods, the Solubilization Index (SI) was computed by getting the ratio of total diameter (colony + halo/clearance) to colony diameter.17 SI of <2.00 is considered low, >2.00-4.00 is intermediate, and >4.00 is high solubilizers.20
Indole-3-acetic acid (IAA) production
Isolates were grown in 5 ml Luria-Bertani (LB) broth tubes supplemented with 100 µg/L tryptophan and incubated at 30 ± 2 °C for 48 hours. Then, 1.5 ml of the broth culture was centrifuged at 10000 rpm for 5 minutes, and the supernatant (approximately 1 ml) was added to an equal volume (1 ml) of Salkowski’s reagent. The contents were mixed and incubated in the dark at room temperature for 30 minutes to develop color. The development of a pink or reddish color indicates IAA production. Absorbance was measured using the UV-Vis spectrophotometer (PG Instruments©, United Kingdom) at 530 nm. Uninoculated broths served as reference solutions. The concentration of the IAA produced was calculated based on the calibration curve of the IAA solutions.21,22
Siderophore (catecholate and hydroxamate) production
Twenty-four-hour bacterial cultures were grown in 10 ml LB broth for 24 hours at 30 ± 2 °C and were centrifuged at 10000 rpm for 5 minutes. For catecholates, 1 ml of bacterial supernatant was mixed with 3 ml of freshly prepared 2% aqueous FeCl3 solution. One milliliter of distilled water and 3 ml of FeCl3 solution were used as a control. For hydroxamates, a pinch of tetrazolium salt was added to the mixture of one to two drops of 2N NaOH and 0.1 ml of the bacterial supernatant. Distilled water was used as a control. The formation of deep red color (presence of hydroxamates) and red wine color (presence of catecholates) was measured using the UV-Vis spectrophotometer (PG Instruments©, United Kingdom) at 431 nm and 495 nm, respectively.23
Gibberellic acid (GA3) production
Bacterial cultures that were grown overnight were centrifuged at 4500 rpm for 5 minutes, then 1 ml of the supernatant was transferred into clear glass tubes, and 1 ml of concentrated HCl and 1 ml of Folin-Ciocalteu reagent were added and brought up to 6 ml with distilled water. The mixtures were placed in the water bath to boil for 5 minutes and then left to cool. The bacterial mixture was prepared in triplicate. The resulting bluish-to-green color was measured at 760 nm using a UV-Vis spectrophotometer (PG Instruments©, United Kingdom). The reading of the bacterial reaction mixtures was averaged, and then the GA3 concentration was estimated based on the calibration curve of GA3.24
Genomic DNA isolation, PCR amplification of the 16S rRNA gene, and sequencing of amplicons
Sequencing was done at Macrogen (Seoul, South Korea). DNA was extracted using the InstaGene Matrix (BIO-RAD, Cat. No. 732-6030) following the kit’s extraction protocols. The quality-checked DNA samples were amplified via PCR by mixing the different components: 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′), Taq buffer, DNA polymerase, and dNTP Mix following the cycling condition 95 °C for 5 minutes, 30 cycles at 95 °C for 1 minute, 60 °C for 45 seconds, 70 °C for 10 minutes, and held at 4 °C. The purified amplicons were subjected to capillary sequencing by incorporation of fluorescently labeled chain terminator ddNTPs. Then, base calling was done using Variant Reporter Software Version 2.1 (Applied Biosystems), DNASTAR Lasergene SeqMan 7.0, and Macrogen SNP analysis program v1.0.
Phylogenetic analysis of the 16S rRNA sequence contigs
The sequence contigs were compared to those in GenBank® of NCBI (www.ncbi.nlm.
nih/gov) using the BLAST program. Using the accession number of each bacterium, their sequences in FASTA format were downloaded, and the phylogenetic tree was constructed using MEGA 11.0.13. The 16S rRNA gene sequences of the bacterial isolates were converted to FASTA format and, together with the sequence file obtained from the BLASTn, were imported into MEGA 11 and were aligned via the MUSCLE tool. After doing the multiple alignment of the nucleotide sequences, the data file was utilized to construct a phylogenetic tree using the Neighbor-Joining Tree method. A Bootstrap method was used with 1000 values to determine the tree’s reliability.25
Bacterial growth determination and rapid detection of CP degradation in liquid medium
Inoculum and media preparation
The isolates were grown in Luria-Bertani broth (100 ml) for 24 hours at 30 ± 2 °C. Cells were harvested via centrifugation at 4600 x g for 5 minutes and washed twice with sterile normal saline solution (NSS). Washed cells were resuspended in sterile NSS and adjusted to an OD of 1.0.
Growth determination and detection of CP biodegradation
Isolates were grown in 100 ml mineral salt medium (MSM) supplemented with 100 mg/L-1 CP at 30 ± 2 °C in a shaker incubator for 7 days. For bacterial growth studies, 1 ml of culture was withdrawn every 24 hours, and growth was measured using a UV-Vis spectrophotometer (PG Instruments™, United Kingdom) at 600 nm. Log10 CFU was calculated using the calibration curve of each isolate.26 The residual CP was determined by collecting 1.5 ml of culture medium and centrifuging at 4 °C at 10000 rpm for 10 minutes. The pellet was discarded, and the absorbance of the cell-free supernatant was determined at 290 nm. The CP concentration was computed using the calibration curve, and the % degradation of each isolate was computed.27 Uninoculated flasks served as the controls.
% CP Biodegradation = [CPinitial – CPfinal / CPfinal ] × 100
Where:
CPinitial – initial concentration of CP calculated based on OD290
CPfinal – final concentration of CP calculated based on OD290
Chlorpyrifos (CP) and malathion (MT) tolerance of soil bacteria
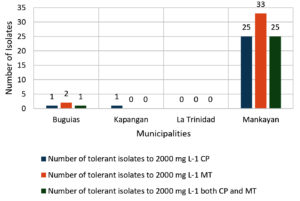
A total of 117 soil bacteria isolated using a 100 mg L-1 starting concentration of chlorpyrifos (CP) and malathion (MT) were culturally and morphologically characterized, and 42 unique isolates were selected and subjected to a tolerance test. The results revealed a remarkable degree of tolerance to high concentrations of CP and MT, as shown in Figure 1. Among the isolates, 27 (64%) showed tolerance of up to 2000 mg L-1 CP, and 35 (83%) were tolerant to the same concentration of MT.
Plant Growth Promoting (PGP) characteristics of soil bacteria
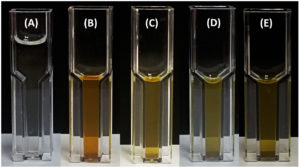
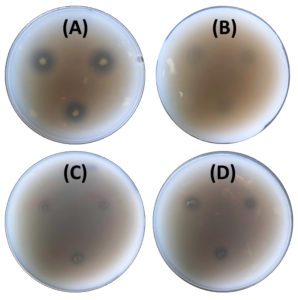
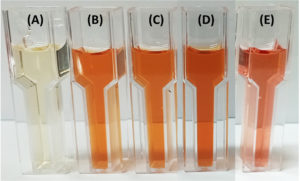
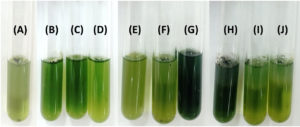
The nitrogen-fixing assay revealed that only four isolates (11%), namely M4C4-5, M4C4-7, M1C4-13, and M1C5-1, exhibited colony growth on nitrogen-free Jensen’s agar medium. High production of ammonia, as shown in the color change of the supernatant, was recorded by M5C4-3 at 116.92 mg L-1 (Figure 2). Only one isolate (M1C4-15) demonstrated phosphate solubilization with an SI of 1.13. Four isolates (M1C4-15, M1C5-1, M1G4-8, and M5C4-13) tested positive for potassium solubilization, with SI values ranging from 1.29 to 2.53, with M1C4-15 showing the highest value of 2.53. This is classified as intermediate based on the literature, while other isolates are classified as low solubilizers (Figure 3). IAA production ranges from 1.29 to 120.86 mg L-1, with the highest production coming from four isolates, namely K2G4-3, B1C5-2, B3C4-6, and M5C4-7, with averages of 120.86, 73.82, 67.50, and 52.21 mg L-1 IAA, respectively. This is based on the intensity of the reddish color as measured by a spectrophotometer (Figure 4). For siderophore detection, results revealed that M5C4-13 produced the highest amount of hydroxamates, with 2.86 au, followed by M1C5-1, with 2.85 au (Figure 5). The lowest detected hydroxamates are 0.32 au by M5C4-3. On the other hand, catecholate production ranges from 0.32 to 0.52 au, with M1G4-8 producing the highest (0.52 au), followed by M1C5-1 (0.51 au). Results showed variability in GA3 production by the soil bacterial isolates, ranging from 38.33 to 79.66 mg L-1. The isolate M5C4-3 exhibited the highest production of GA3 (79.66 mg L-1), followed by K2G4-3 (75.10 mg L-1) (Figure 6).
Figure 2. Ammonia production by bacterial isolates following reaction with Nessler’s reagent. Culture supernatants were mixed with Nessler’s reagent, and the resulting color change was assessed in cuvettes: (A) control (uninoculated medium), (B) M5C4-3, (C) M1C5-1, (D) M4C4-5, and (E) M4C4-7. Development of a yellow to brown coloration is indicative of ammonia production
Figure 3. Potassium solubilization activity of bacterial isolates on Aleksandrov agar. Clear zones surrounding the colonies indicate potassium solubilization. The positive isolates are: (A) M1C4-15, (B) M1C5-1, (C) M5C4-13, and (D) M1G4-8
Figure 4. Indole-3-acetic acid (IAA) production by bacterial isolates following the addition of Salkowski reagent. Development of pink coloration indicates IAA production. The treatments were: (A) control (uninoculated medium), (B) B1C5-2, (C) B3C4-6, (D) K2G4-3, and (E) M5C4-7
Figure 5. Siderophore production by bacterial isolates. Hydroxamate-type siderophores were indicated by the development of an intense deep red color (A), while catecholate-type siderophores exhibited a red wine to brown coloration (B). Tubes: (1) control (uninoculated medium), (2) B1C5-2, (3) K2G4-3, (4) M5C4-13, (5) M1G4-8, (6) M1C4-15, (7) M4C4-7, (8) M5C4-3, (9) M1C5-1, and (10) M4C4-5
Figure 6. Gibberellic acid (GA₃) production by bacterial isolates. Culture supernatants were treated with HCl and Folin-Ciocalteu reagents, and the development of a darker coloration indicated higher GA₃ production. Tubes: (A) control (uninoculated medium), (B) M4C4-5, (C) M5C4-3, (D) M1G4-8, (E) M4C4-7, (F) M5C4-13, (G) K2G4-3, (H) B1C5-2, (I) M1C5-1, and (J) M1C4-13
Molecular identification of the chlorpyrifos-malathion – tolerant and PGP isolates
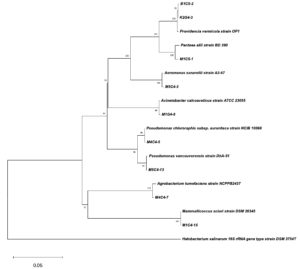
The nine OP-tolerant and PGP bacterial isolates were identified by 16S rRNA sequencing as Pseudomonas chlororaphis (M4C4-5), Pantoea allii (M1C5-1), Aeromonas sanarellii (M5C4-3), Agrobacterium tumefaciens (M4C4-7), Mammaliicoccus sciuri (M1C4-15), Acinetobacter calcoaceticus (M1G4-8), Pseudomonas vancouverensis (M5C4-13), and Providencia vermicola (K2G4-3/B1C5-2) with percent similarities ranging from 98.77 to 100%. The bootstrap tree is shown in Figure 7. Table 1 summarizes the PGP traits of the identified bacterial isolates.
Table (1):
PGP characteristics of the identified bacterial isolates
| Isolates | NF | AP mg L-1 | PS (SI) | KS (SI) | IAA | SH | SC | GA |
|---|---|---|---|---|---|---|---|---|
| mg L-1 | au | au | mg L-1 | |||||
| P. chlororaphis | + | 105.0 ± 0.1 | – | – | 29.3 ± 1.6 | 0.5 | 2.5 | 61.2 ± 0.0 |
| P. allii | + | 105.9 ± 0.0 | – | 1.3 ± 0.0 | 19.0 ± 0.1 | 0.5 | 2.9 | 51.8 ± 0.0 |
| A. sanarellii | + | 116.9 ± 0.1 | – | – | 8.0 ± 0.1 | 0.3 | 0.3 | 79.7 ± 0.0 |
| A. tumefaciens | + | 105.9 ± 0.1 | – | – | 9.1 ± 0.1 | 0.4 | 1.7 | 44.7 ± 0.0 |
| M. sciuri | – | – | 1.1 ± 0.0 | 2.5 ± 0.1 | 34.6 ± 0.2 | 0.4 | 0.5 | 51.6 ± 0.0 |
| A. calcoaceticus | – | – | – | 1.7 ± 0.3 | 15.5 ± 0.1 | 0.5 | 1.6 | 49.2 ± 0.0 |
| P. vancouverensis | – | – | – | 1.6 ± 0.3 | 19.8 ± 0.4 | 0.4 | 2.9 | 54.6 ± 0.0 |
| P. vermicola | – | – | – | – | 120.9 ± 2.1 | 0.4 | 0.9 | 75.1 ± 0.0 |
| P. vermicola | – | – | – | – | 73.8 ± 1.3 | 0.4 | 0.9 | 38.3 ± 0.0 |
NF = Nitrogen Fixation (+/-); AP = Ammonia Prod (mg L-1); PS = Phosphate Solubilization (Solubilization Index); KS = Potassium Solubilization (Solubilization Index); IAA = Indole Acetic Acid Production (mg L-1); SH = Siderophore – Hydroxamate Production (absorbance unit); SC = Siderophore – Catecholate Production (absorbance unit); GA = Gibberellic Acid Production (mg L-1)
Figure 7. Phylogenetic tree based on 16S rRNA gene sequences showing the relationship between isolated bacterial strains (labeled with alphanumeric codes, e.g., B1C5-2, M1G4-8) and reference strains from the NCBI database. The tree was constructed using the Neighbor-Joining method, and bootstrap values (expressed as percentages of 1000 replicates) greater than 50% are shown at the nodes, indicating the confidence level of each branch. Halobacterium salinarum DSM 3754T was used as the outgroup to root the tree. The scale bar represents 0.05 nucleotide substitutions per site
Bacterial growth analysis and chlorpyrifos degradation in liquid media
Three bacterial isolates that are classified under biosafety level 1 based on Leibniz Institute – German Collection of Microorganisms and Cell Cultures (DSMZ), American Biological Safety Association (ABSA), American Type Culture Collection (ATCC), and ePATHogen Risk Group Database of Canada were subjected to a pesticide biodegradation assay (Table 2). The bacterial growth and detection of the biodegradation ability of the bacterial isolates in a liquid medium containing CP were determined by UV-Vis spectrophotometry (PG Instruments©, United Kingdom).
Table (2):
The biosafety levels of the bacterial isolates were identified
Isolates |
DSMZ |
ABSA |
ATCC |
ePATHogen |
|---|---|---|---|---|
P. chlororaphis |
1 |
NRG |
1 |
RG1a, RG1h |
P. allii |
1 |
NRG |
1 |
RG1a, RG1h |
A. sanarellii |
2 |
2 (hp) |
2 |
RG2h, RG1a |
A. tumefaciens |
1 ** |
NRG |
1 |
RG1a, RG1h |
M. sciuri |
NRG |
NRG |
1 |
RG1a, RG1h |
A. calcoaceticus |
2 |
2 |
2 |
RG1/2h, RG2a |
P. vancouverensis |
1 |
NRG |
1 |
RG1a, RG1h |
P. vermicola |
NRG |
NRG |
NRG |
NRG but other genus are RG1/2h, RG1a |
1: Microbes that are not known to consistently cause disease in healthy adults and present minimal potential hazard to laboratorians and the environment; 2: Microbes there pose moderate hazards to laboratory workers and the environment; ** = known plant pathogen; NRG = no risk group records; ap = animal pathogen; hp = human pathogen; RG1/2a = risk group 1 or 2 for animal; RG1/2h = risk group 1 or 2 for human
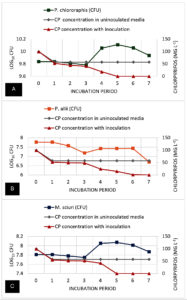
Figure 8. Bacterial growth (cfu) and chlorpyrifos (CP) pesticide residue concentration (mg L-1) dynamics during the 8-day degradation experiment. A) P. chlororaphis; B) P. allii; and C) M. sciuri
Figure 8 shows the growth curve and the chlorpyrifos degradation over seven days. The growth curve shows that the uninoculated control with no pesticide remained relatively stable across the 7-day incubation period. Similarly, Treatment 2, containing 100 ppm CP without inoculation, showed no fluctuations in OD values (0.07) over the incubation period, suggesting that no other factors (i.e., photodegradation, volatilization) affected the turbidity or CP concentration of the MSM. However, Treatment 3, where P. chlororaphis was added, showed significant changes in their CFU values from day 1 to day 3 (9.84 to 9.78 log10 CFU) and then gradually increased on day 4 (10.06 log10 CFU) before peaking at day 5 (10.11 log10 CFU). This was followed by a minor decline to 10.06 log10 CFU on day 6 and 9.94 log10 CFU on day 7. In terms of bacterial growth of P. allii, it was observed that there was a consistent decline in its CFU values over the incubation period. On days 1, 2, and 3, a significant decrease was noticeable, similar to P. chlororaphis from 7.77 log10 CFU to 7.18 log10 CFU, suggesting that the presence of CP in the media may also affect the ability of the bacteria to divide or, in other cases, destroy the cell. However, on day 4, a slight increase in the CFU suggests a minimal growth of the isolate, and then it entered a stationary phase on day 5. After this, a gradual decline in the CFU on days 6 and 7 (7.43 log10 CFU and 6.71 log10 CFU) suggests a significant decline in bacterial cells. Moreover, M. sciuri also showed an abrupt decrease in CFU values (7.81 log10 CFU to 7.75 log10 CFU) in just three days, but peaked on day 4 (8.05 log10 CFU). The CFU values continued to increase on day 5 and then gradually decreased on days 6 and 7 (8.01 log10 CFU and 7.87 log10 CFU), implying that bacterial growth stopped and started declining, which may be due to the waste by-products of the bacterial cells.
No changes in CP concentration were seen after days 1 to 7 in the CP-amended media without inoculants (T2). Although there was as much as 60% (43 mg L-1) reduction in the initial CP concentration within 24 hours for all treatments (T2, T3, and T4). P. chlororaphis-treated media (T3) showed biodegradation from day 1, with complete degradation achieved at day 5. Significantly, the highest degradation was observed between days 4 to 5, with the sharpest decrease in CP level from 18.24 to 0 mg L-1. P. allii showed a slower rate of biodegradation compared to P. chlororaphis, with a steady decrease in CP concentration observed from day 1 to 6 and complete degradation on day 7. Similar to P. chlororaphis, M. sciuri was able to degrade 100% of the amended CP in the media within 5 days. A gradual decline in the CP concentration was observed from days 1 to 4, followed by a sudden decrease on day 5, with 100% degradation of amended CP. In summary, P. chlororaphis and M. sciuri were able to achieve 100% degradation of CP in 5 days; however, P. chlororaphis efficiently degraded most of the amended CP by day 4 (81.76%) compared to M. sciuri (58.26%). On the other hand, P. allii appears to be the least efficient degrader of Treatment 3. Additionally, the decline in P. allii population at day 3 suggests brief toxicity of the pesticide to the bacterial population before adaptation and degradation are achieved.
The high number of CP- and MT-tolerant bacterial isolates obtained from vegetable farms in Benguet Province suggests that soil bacterial populations in this region possess or have developed structural and/or physiological mechanisms allowing them to survive the toxicity of these organophosphate residues and even metabolically break these down for carbon sources. The introduction of pesticides into their environment triggers a defensive adaptation in certain microbes, including the formation of biofilms, induced mutations, horizontal and vertical gene transfer, and increased expression of certain hydrolytic enzymes.28 The observed difference in bacterial resistance and degradation capabilities between MT and CP could be attributed to the chemical structure and toxicity of the pesticides, which may influence the type of bacterial enzymes and metabolic pathways activated. Furthermore, 3,5,6-trichloro-2-pyridinol (TCP), the hydrolytic product of CP, contains antibacterial properties and inhibitory effects that may interfere with growth processes.29 A 200 mg L-1 of TCP exerted more toxicity toward bacterial isolates by inhibiting their growth and colony formation within 48 hours.30 Moreover, plasmids encoding degradation pathways, as seen in a Burkholderia cepacia group strain that degraded chlorpyrifos-methyl, suggest a genetic basis for resistance.31 Overall, the development of bacterial resistance against MT and CP is a complex process influenced by the specific bacterial strain, the presence of genetic elements such as plasmids, and the ecological context. While some bacteria have evolved to degrade MT more widely and efficiently, others have developed specific mechanisms to resist and degrade CP, albeit in fewer numbers.32-34
Three bacterial strains, namely P. chlororaphis, P. allii, and M. sciuri were found to be CP-degraders with plant growth-promoting (PGP) properties, representing a first-hand report of such activity in these species under chlorpyrifos stress. Based on the growth and biodegradation analysis, their growth patterns (both OD600 and CFU values) revealed that these bacteria do not immediately utilize the supplemented CP, which may be attributed to pesticide-induced inhibition of some essential proteins needed for cell division.35 However, once the bacteria acclimatized, they were able to adapt and utilize the CP, thus initiating the renewed proliferation, followed by a decline, usually attributed to depletion of nutrients and build-up of metabolic by-products. These can include CP intermediates, such as TCP,36 acidification of the media due to production of organic acids such as phosphorothioic acid, maleic acid, pyruvic acid, and carboxylic acids,37 and accumulation of waste products.15 The strain P. chlororaphis was able to remove 100% of the initial 100 mg L-1 of CP, without additional carbon source, within just 5 days, suggesting it has higher potential for CP degradation among the three strains tested. In comparison to P. fluorescens, which was previously found to degrade 78.19% of 500 mg L-1 CP within 15 days, P. chlororaphis appears as a strong candidate for bioremediating pesticide-contaminated soils.38 Other species of Pseudomonas have also been reported to degrade as much as 74.60% of the amended CP in MSM within 5 days.39 P. aeruginosa can degrade CP in a liquid medium within 48 hours of incubation with a degradation efficiency of 71%. In addition, P. chlororaphis exhibited certain PGP traits that help in the CP degradation, like high production of EPS that allows bacteria to form biofilms and act as a protective barrier against pesticides and other stresses.16,40,41 In addition, it was reported that S. aureus was able to degrade 1000 mg L-1 CP in MSM.42
The ability of the isolated CP-degrading bacteria to fix nitrogen may be due to the production of nitrogenase, which catalyzes the biological reduction of atmospheric nitrogen shown on Jensen’s agar, and further by positive ammonia production.43,44 Nitrogen fixation carried out by these microorganisms plays an important role in the processes of the nitrogen cycle and its availability for plant uptake.45 Soil bacteria like Bacillus sp., Citrobacter sp., Nitratireductor lucknowense, Pseudomonas nitroreducens, Pseudomonas putida, and Paracoccus sp. NITDBR1 isolated from pesticide-contaminated agricultural soil is among other reported nitrogen-fixing bacteria with pesticide-degrading abilities.46-49 Among the isolates, M. sciuri, demonstrated both phosphate and potassium solubilization abilities, albeit, low for phosphate solubilization. The inorganic phosphate in agar medium is often in a form that is insoluble or not readily available for uptake by bacteria because not all bacteria possess the necessary mechanisms to solubilize, such as the production of organic acids (gluconic and malic) and convert these insoluble phosphate compounds into a form that they can assimilate like in the case of phosphate solubilizing bacterial (PSB) species of Bacillus and Pseudomonas.50-52 The efficiency of PSBs in solubilizing phosphate has been demonstrated in various studies, showing their potential to enhance plant growth and development by increasing the availability of phosphorus, a key nutrient limiting in many soils.30,53,54 In contrast, the high solubilizing activity of M. sciuri to potassium may be due to mechanisms such as secretion of organic acids and enzymes, resulting in the lowering of the pH and breaking down these minerals into plant-accessible forms.55,56 The bacterial strains also produced IAA ranging from 19-35 mg L-1, which is a critical factor in PGP, despite being isolated from pesticide-contaminated soils. Various researchers have also identified other soil bacteria capable of producing IAA, even under the stress of pesticide contamination. These include Priestia megaterium NRRU-BW3, Bacillus siamensis NRRU-BW9, and Bacillus amyloliquefaciens NRRU-TV11, which have been shown to degrade CP while producing IAA, demonstrating their potential in bioremediation and plant growth improvement in contaminated agricultural soils.57 Similarly, Achromobacter spanius and Serratia plymuthica, isolated from Medicago sativa nodules, have shown the ability to produce IAA and enhance alfalfa dry weight, indicating their role in phytostabilizing pesticide-contaminated soils.26 On top of other PGP traits, these bacteria represent promising candidates for biofertilizer development. The application of such IAA-producing bacteria could enhance root growth and development, improve nutrient uptake, and increase stress tolerance in crops.51,58-60 Moreover, the bacterial isolates also demonstrated the production of siderophores and gibberellic acids. Siderophores are iron chelators due to their high affinity for ferric ions. This metal is crucial for the growth and development of nearly all living organisms. It catalyzes essential enzymatic processes, such as oxygen metabolism, electron transport, and the creation of DNA and RNA.61 Not only that, it was reported that through the production of siderophores, microorganisms are protected from metal toxicities. The strains P. chlororaphis and P. allii were able to produce siderophores such as hydroxamates, ranging from 0.46-0.51 au and catecholates, ranging from 2.52-2.85 au. Other species, like P. aeruginosa, were reported to produce siderophores that can sequester other metals in their environment and prevent them from diffusing across the cell membranes.62 Similarly, P. fluorescens was also found to produce pyoverdine siderophore that increases mobility and reduces the toxicity of heavy metals in uranium mines. P. aeruginosa and P. fluorescens were further shown to increase the rate of phytoextraction and phytoremediation of heavy metals.63,64 Finally, P. chlororaphis, P. allii, and M. sciuri can promote crop growth under pesticide stress through GA3 production, which was shown to range from 51-61 mg L-1. GA3 has an important role in stem and leaf expansion, seed germination, and an increase in the number of root hairs.65,66 Various authors have reported that species of soil bacteria, such as Acetobacter, Azobacter, Azospirillum, Micrococcus sp., Staphylococcus, Bacillus, and Pseudomonas, can synthesize GA3.67,68 For instance, B. licheniformis and B. pumilus have a positive effect on Pinus pinea by increasing leaf and root length.69 Similarly, Rhizobium strains promote root growth in rice by the production of GA3 and IAA.70
The results of this study showed that P. chlororaphis, P. allii, and M. sciuri from vegetable farms in Benguet are excellent candidates for the development of microbial inoculum to reduce pesticide residues in farm soils and crops. Furthermore, their ability to promote plant growth through nitrogen fixation, ammonia production, potassium and phosphate solubilizations, and production of indole acetic acid, siderophores, and gibberellic acids provides an added advantage in the development of formulations for enhancing crop performance under stressful nutrient-deficient conditions. Thus, these data provide new insights and possible applications in bioremediation and sustainable agriculture.
ACKNOWLEDGMENTS
The authors would like to acknowledge the Department of Science and Technology (DOST) Grant-In-Aid (GIA) through the Philippine Council for Agriculture and Aquatic Resources Research and Development (PCAARRD) for funding the research project under the Grant-in-Aid (GIA) program, which supported this study. Also, the authors are grateful to the Graduate Research Education Assistantship for Technology (GREAT) Program of DOST for supporting the academic endeavors of the primary author.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
FUNDING
This study was funded by the GREAT Program and the DOST-PCAARRD GIA program.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Chaud M, Souto EB, Zielinska A, et al. Nanopesticides in agriculture: benefits and challenges in agricultural productivity, toxicological risks to human health and environment. Toxics. 2021;9(6):131.
Crossref - Food and Agriculture Organization (FOA). Pesticide use, pesticide trade and pesticide indicators. Global, regional and country trends. 1990-2020. 2022
Crossref - World Health Organization (WHO). Pesticide residues in food: Report of the Joint Meeting of the FAO Panel Experts on Pesticide Residues in Food and the Environment and the WHO Core Assessment Group. FAO Plant Production and Protection. 2022;163.
- Song F, Swinton SM. Returns to integrated pest management research and outreach for soybean aphid. J Econ Entomol. 2009;102(6):2116-2125.
Crossref - Cid AP, del Mundo FR, Espino MPB. A modified analytical procedure for the determination of carbaryl, carbofuran and methomyl residues in agricultural soil and river water samples from La Trinidad, Benguet and Aurora, Isabela, Philippines. Philipp Agric Sci. 2006;89(1):71-84.
- Lurean CP, Lucas CP, Kisim DD. Soil properties of agricultural farms in two agro-ecological zones of three municipalities of Benguet. Mt J Sci Interdiscip Res. 2016;75:16-29.
Crossref - Reyes G, Gomez R Jr, Fomeg-as D, Lasangen WC, Anacin C. Detection of organophosphate residues in selected crops in Benguet and Mt. Province, Philippines. J Environ Sci Manag. 2017;20(2):28-33.
Crossref - Lu JLDP. Multipesticide residue assessment of agricultural soil and water in major farming areas in Benguet, Philippines. Arch Environ Contam Toxicol. 2010;59(2):175-181.
Crossref - Sehrawat A, Phour M, Kumar R, Sindhu SS. Bioremediation of Pesticides: An Eco-Friendly Approach for Environment Sustainability. In: Panpatte, DG, Jhala, YK. (eds) Microbial Rejuvenation of Polluted Environment. Microorganisms for Sustainability, vol 25. Springer, Singapore. 2021;1:23-84.
Crossref - Aswathi A, Pandey A, Madhavan A, Sukumaran RK. Chlorpyrifos induced proteome remodelling of Pseudomonas nitroreducens AR-3 potentially aid efficient degradation of the pesticide. Environ Technol Innov. 2021;21:101307.
Crossref - Huang Y, Zhang W, Pang S, et al. Insights into the microbial degradation and catalytic mechanisms of chlorpyrifos. Environ Res. 2021;194:110660.
Crossref - Akbar S, Sultan S. Soil bacteria showing a potential of chlorpyrifos degradation and plant growth enhancement. Braz J Microbiol. 2016;47(3):563-570.
Crossref - Kaur M, Vyas P, Rahi P, Sharma S. Chlorpyrifos-and carbofuran-tolerant phosphate-solubilising Arthrobacter oxydans and Bacillus flexus improved growth and phosphorus content in potato in pesticide-amended soils. Potato Res. 2022;65(2):213-231.
Crossref - Zhao L, Wang F, Zhao J. Identification and functional characteristics of chlorpyrifos degrading and plant growth-promoting bacterium Acinetobacter calcoaceticus. J Basic Microbiol. 2014;54(5):457-463.
Crossref - Simon-Delso N, Amaral-Rogers V, Belzunces LP, et al. Systemic insecticides (neonicotinoids and fipronil): trends, uses, mode of action, and metabolites. Environ Sci Pollut Res. 2015;22:5-34.
Crossref - Naseem M, Chaudhry AN, Jilani G, et al. Exopolysaccharide-producing bacterial cultures of Bacillus cereus and Pseudomonas aeruginosa in soil augment water retention and maize growth. Heliyon. 2024;10(4):e26104.
Crossref - Etesami H, Emami S, Alikhani HA. Potassium solubilizing bacteria (KSB): Mechanisms, promotion of plant growth, and prospects-a review. J Soil Sci Plant Nutr. 2017;17(4):897-911.
Crossref - Shomi FY, Uddin MB, Zerin T. Isolation and characterization of nitrogen-fixing bacteria from soil sample in Dhaka, Bangladesh. Stamford J Microbiol. 2021;11(1):11-13.
Crossref - Abdelwahed S, Trabelsi E, Saadouli I, et al. A new pioneer colorimetric micro-plate method for the estimation of ammonia production by plant growth-promoting rhizobacteria (PGPR). Main Group Chem. 2022;21(1):55-68.
Crossref - Saha M, Maurya BR, Meena VS, Bahadur I, Kumar A. Identification and characterization of potassium solubilizing bacteria (KSB) from Indo-Gangetic Plains of India. Biocatal Agric Biotechnol. 2016;7:202-209.
Crossref - Sulistya IR, Sitoresmi P, Agung W, Rina TS, Achmad R. Quantitative assay of indole acetic acid-producing bacteria isolated from several lakes in East Java, Indonesia. Biodiversitas J Biol Divers. 2020;21(11).
Crossref - Shoukry AA, El-Sebaay HH, El-Ghomary AE. Assessment of indole acetic acid production from Rhizobium leguminosarum strains. Curr Sci Int. 2018;7(1):60-69.
- Carson KC, Holliday S, Glenn AR, Dilworth MJ. Siderophore and organic acid production in root nodule bacteria. Arch Microbiol. 1992;157(3):264-271.
Crossref - Abou-Aly HE, Youssef AM, El-Meihy RM, Tawfik TA, El-Akshar EA. Evaluation of heavy metals tolerant bacterial strains as antioxidant agents and plant growth promoters. Biocatal Agric Biotechnol. 2019;19:101110.
Crossref - Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
- Saengsanga T, Phakratok N. Biodegradation of chlorpyrifos by soil bacteria and their effects on growth of rice seedlings under pesticide-contaminated soil. Plant Soil Environ. 2023;69(5):210-220.
Crossref - Hamsavathani V, Aysha OS, Ajith AR. Isolation and identification of chlorpyrifos degrading bacteria from agricultural soil. Int J Adv Res. 2017;5(5):1209-1221.
Crossref - Rangasamy K, Athiappan M, Devarajan N, et al. Pesticide-degrading natural multidrug-resistant bacterial flora. Microb Pathog. 2018;114:304-310.
Crossref - Yadav M, Shukla AK, Srivastva N, Upadhyay SN, Dubey SK. Utilization of microbial community potential for removal of chlorpyrifos: a review. Crit Rev Biotechnol. 2016;36(4):727-742.
Crossref - Bhardwaj S, Kaushal R, Jhilta P, Rana A, Dipta B. Phosphate Solubilizing Microorganisms: Potential Bioinoculants for Sustainable Agriculture. In: Prasad, R., Zhang, SH. (eds) Beneficial Microorganisms in Agriculture. Environmental and Microbial Biotechnology. Springer, Singapore. 2022:131-159.
Crossref - Ahmed F, Azmy AE, Saafan TM, Essam MA, Amin S, Shaban HA. Biodegradation of Malathion by Acinetobacter baumannii strain AFA isolated from domestic sewage in Egypt. World Acad Sci Eng Technol Int J Biol Biomol Agric Food Biotechnol Eng. 2015;9(1):55-65.
- Isworo S, Oetari PS. The chemical compounds from degradation of profenofos and malathion by indigenous bacterial consortium. J Pure Appl Microbiol. 2021;15(2):897-914.
Crossref - Kim JR, Ahn YJ. Identification and characterization of chlorpyrifos-methyl and 3,5,6-trichloro-2-pyridinol degrading Burkholderia sp. strain KR100. Biodegradation. 2009;20(4):487-497.
Crossref - Ajaz M, Jabeen N, Akhtar S, Rasool SA. Chlorpyrifos resistant bacteria from Pakistani soils: isolation, identification, resistance profile and growth kinetics. 2005. Pakistan Journal of Botany, 37(2):381.
- Ambreen S, Yasmin A. Novel degradation pathways for Chlorpyrifos and 3, 5, 6-Trichloro-2-pyridinol degradation by bacterial strain Bacillus thuringiensis MB497 isolated from agricultural fields of Mianwali, Pakistan. Pestic Biochem Physiol. 2021;172:104750.
Crossref - Nandhini AR, Harshiny M, Gummadi SN. Chlorpyrifos in environment and food: a critical review of detection methods and degradation pathways. Environ Sci: Processes Impacts. 2021;23(9):1255-1277.
Crossref - Fernandez-Martinez LT, Javelle A, Hoskisson PA. Microbial primer: bacterial growth kinetics. Microbiology. 2024;170(2):001428.
Crossref - Dubey S, Dhanya MS. Chlorpyrifos degradation in semi-arid soil by Pseudomonas fluorescens strain CD5 isolated from manured soil. Soil Sediment Contam. 2023;32(4):460-477.
Crossref - Zhang Q, Li S, Ma C, Wu N, Li C, Yang X. Simultaneous biodegradation of bifenthrin and chlorpyrifos by Pseudomonas sp. CB2. J Environ Sci Health B. 2018;53(5):304-312.
Crossref - Bhatia D, Malik DK. Isolation and characterization of chlorpyrifos degrading soil bacteria of environmental and agronomic significance. J Environ Sci Eng. 2013;55(2):227-238.
- Vu B, Chen M, Crawford RJ, Ivanova EP. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules. 2009;14(7):2535-2554.
Crossref - Oruc HH. Fungicides and their effects on animals. In: Carisse O, ed. Fungicides. In-Tech Publishers; 2010:349-362.
Crossref - Davis KE, Joseph SJ, Janssen PH. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl Environ Microbiol. 2005;71(2):826-834.
Crossref - Berman-Frank I, Lundgren P, Falkowski P. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res Microbiol. 2003;154(3):157-164.
Crossref - Vance CP, Graham PH, Allan DL. Biological Nitrogen Fixation: Phosphorus – A Critical Future Need?. In: Pedrosa FO, Hungria M, Yates G, Newton WE. (eds) Nitrogen Fixation: From Molecules to Crop Productivity. Current Plant Science and Biotechnology in Agriculture, vol 38. Springer, Dordrecht. 2000:509-514.
Crossref - Kapta S, Madathil V, Kalola M, Kothari RK, Rank J. Preparation of chlorpyrifos degradation and plant growth-promoting bacterial formulation. SSRN Electronic Journal. 2020.
Crossref - Sahoo B, Ningthoujam R, Chaudhuri S. Isolation and characterization of a lindane-degrading bacterium Paracoccus sp. NITDBR1 and evaluation of its plant growth-promoting traits. Int Microbiol. 2019;22(1):155-167.
Crossref - Sahoo B, Chaudhuri S. Screening of lindane-degrading bacteria isolated from soil for their plant growth-promoting attributes. Environ Sustain. 2019;2(2):97-106.
Crossref - Bhadbhade BJ, Sarnaik SS, Kanekar PP. Biomineralization of an organophosphorus pesticide, Monocrotophos, by soil bacteria. J Appl Microbiol. 2002;93(2):224-234.
Crossref - Yu H, Wu X, Zhang G, et al. Identification of the phosphorus-solubilizing bacteria strain JP233 and its effects on soil phosphorus leaching loss and crop growth. Front Microbiol. 2022;13:892533.
Crossref - Timofeeva A, Galyamova M, Sedykh S. Prospects for using phosphate-solubilizing microorganisms as natural fertilizers in agriculture. Plants. 2022;11(16):2119.
Crossref - Sanchez-Gonzalez ME, Mora-Herrera ME, Wong-Villarreal A, et al. Effect of pH and carbon source on phosphate solubilization by bacterial strains in Pikovskaya medium. Microorganisms. 2022;11(1):49.
Crossref - Ahmad A, Moin SF, Liaqat L, et al. Isolation, solubilization of inorganic phosphate, and production of organic acids by individual and co-inoculated microorganisms. Geomicrobiol J. 2023;40(1):111-121.
Crossref - Aliyat FZ, Maldani M, El Guilli M, Nassiri L, Ibubijen J. Phosphate-solubilizing bacteria isolated from phosphate solid sludge and their ability to solubilize three inorganic phosphate forms: Calcium, iron, and aluminum phosphates. Microorganisms. 2022;10(5):980.
Crossref - Sood Y, Singhmar R, Singh V, Malik DK. Isolation and characterization of potential potassium solubilizing bacteria with various plant growth-promoting traits. Biosci Biotechnol Res Asia. 2023;20(1):79-84.
Crossref - Berde CV, Gawde SS, Berde VB. Potassium Solubilization: Mechanism and Functional Impact on Plant Growth. In: Yadav, A.N. (eds) Soil Microbiomes for Sustainable Agriculture. Sustainable Development and Biodiversity, vol 27. Springer, Cham. 2021:133-148.
Crossref - Raiawat MVS, Ansari WA, Singh D, Singh R. Potassium Solubilizing Bacteria (KSB). In: Singh, D., Prabha, R. (eds) Microbial Interventions in Agriculture and Environment. Springer, Singapore. 2019:189-209.
Crossref - Aroua I, Abid G, Souissi F, et al. Identification of two pesticide-tolerant bacteria isolated from Medicago sativa nodule useful for organic soil phytostabilization. Int Microbiol. 2019;22(1):111-120.
Crossref - Egamberdieva D, Wirth SJ, Alqarawi AA, Abd_Allah EF, Hashem A. Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front Microbiol. 2017;8:278255.
Crossref - Vessey JK. Plant growth-promoting rhizobacteria as biofertilizers. Plant Soil. 2003;255(2):571-586.
Crossref - Chagas Junior AF, de Oliveira AG, de Oliveira LA, et al. Production of indole-3-acetic acid by Bacillus isolated from different soils. Bulg J Agric Sci. 2015;21(2):282-287.
- Schalk IJ, Hannauer M, Braud A. New roles for bacterial siderophores in metal transport and tolerance. Environ Microbiol. 2011;13(11):2844-2854.
Crossref - Edberg F, Kalinowski BE, Holmstrom SJM, Holm K. Mobilization of metals from uranium mine waste: the role of pyoverdines produced by Pseudomonas fluorescens. Geobiology. 2010;8(4):278-292.
Crossref - Braud A, Jezequel K, Bazot S, Lebeau T. Enhanced phytoextraction of an agricultural Cr- and Pb-contaminated soil by bioaugmentation with siderophore-producing bacteria. Chemosphere. 2009;74(2):280-286.
Crossref - Castro-Camba R, Sanchez C, Vidal N, Vielba JM. Plant development and crop yield: The role of gibberellins. Plants. 2022;11(19):2650.
Crossref - Passari AK, Mishra VK, Leo VV, Gupta VK, Singh BP. Phytohormone production endowed with antagonistic potential and plant growth-promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiol Res. 2016;193:57-73.
Crossref - Leslie B, Scheila D, Madhumitta D, et al. Exporting harm: the high-tech trashing of Asia. The Basel Action Network, Seattle. 2002.
- Mansour FA, Aldesuquy HS. Studies on plant growth regulators and enzyme production by some bacteria. Qatar Univ Sci J. 2014;159:121-128.
- Probanza A, Garcia JAL, Palomino MR, Ramos B, Manero FJG. Pinus pinea L. seedling growth and bacterial rhizosphere structure after inoculation with PGPR Bacillus (B. licheniformis CECT 5106 and B. pumilus CECT 5105). Appl Soil Ecol. 2002;20(2):75-84.
Crossref - Yanni YG, Rizk RY, Abd El-Fattah FK, et al. The beneficial plant growth-promoting association of Rhizobium leguminosarum bv. trifolii with rice roots. Funct Plant Biol. 2001;28(9):845-870.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.