ISSN: 0973-7510
E-ISSN: 2581-690X
Rotavirus A (RVA) causes viral gastroenteritis in humans and animals, including calves, piglets, and foals. The current study reports the genetic characterization of the full-length enterotoxin gene, NSP4, from caprine and ovine species. Upon characterizing eight full-length NSP4 genes by sequencing, it was found that the four caprine and three ovine RVAs NSP4 genes are of E2 genotype and the sole ovine RVA isolate was found to be of E1 genotype. In the sequence and phyloanalysis of the NSP4 gene the seven E2 genotypes clustered with bovine, human, and caprine isolates from India and Bangladesh, respectively. The E1 genotype of ovine RVA was closer to human RVA isolate from India. The nucleotide per cent identity analysis revealed that all E2 genotype strains of caprine and ovine species ranged from 88.4% to 90.4% and it was found common to both the reference human RVA isolates DS-1 and AU-1. Whereas, the E1 genotype ovine strain clustered with human RVA isolates with 93.1% nucleotide per cent identity. The RVA strains circulating in caprine and ovine populations may share a common origin which is usually found in artiodactyl species because humans share a common dwelling with animals. Future studies are needed to confirm these findings of their relationship with humans and large animals.
Rotavirus, caprine, ovine, NSP4, enterotoxin, sequencing, phylogenetic analysis
Rotavirus A (RVA) is among the leading causes of virus mediated gastroenteritis in humans and animals worldwide1. In livestock, RVA causes severe enteric infection in calves, piglets, and foals2. RVAs belong to the family Reoviridae which possesses 11 dsRNA segmented genome which encodes for six structural (VP1-4 and VP6-7) and six non-structural proteins (NSP1-6). On the basis of group-specific capsid protein VP6 gene, RVA is classified into nine species, namely A-I (RVA, RVB….RVI). Of them, three species (RVA, RVB, & RVC) have been reported in small ruminants3. RVAs have been studied extensively for domesticated species like bovine, porcine and humans, but caprine and ovine species have remained neglected4. Apart from RVs other enteric viruses are being discovered and studied which are also associated with diarrhoeal problems in small ruminants5-7.
Among the different proteins of rotaviruses, NSP4 plays the role of viral enterotoxin, which has been described to induce diarrhoea in suckling mice8. This discovery led to an increase in scientific interest to understand the role of the NSP4 enterotoxin gene in inducing diarrhoea in different animals and human neonates. NSP4, consisting of 175 amino acids, is an endoplasmic reticulum (ER) specific glycoprotein that has a role in viral pathogenesis and morphogenesis9. It has three hydrophobic domains, viz – H1 domain (7-21 aa), H2 domain (29-47 aa), and H3 domain (67-85 aa), and a coiled alpha helix domain stretches from 95-137 aa1. The amino-terminus region is located from 1-44 aa inside the lumen of ER, whereas 45-175 aa stretch towards carboxyl-terminus contains the cytoplasmic tail (CT) known for key biological functions related to the protein, which includes a variation of Ca2+ homeostasis by discharging Ca2+ from the ER10-12, Ca2+ and VP4 binding1, permeabilization into the plasma membrane13, recruiting the double-layered particle to the lumen of ER to be matured into triple-layered particle14,15, and induction of diarrhoea in neonatal mice8,16,17.
So far, 27 E genotypes (E1-E27) have been identified in different animal and human species established on a 85% identity cut-off value for the NSP4 gene18,19. Genotypes E1, E2, and E3 have been reported more commonly18. Usually, all the NSP4 genotypes tend to segregate according to the RVA host species20 suggesting the dominance of specific genotypes for a particular species. To date, the RVA NSP4 and other genes of RVs have been analysed in more detail for human21-24, bovine25, porcine26, and avian species27. However, small ruminants remain the least studied species worldwide. RVA infection has been reported from all over the world, but reports from India are scanty, especially from caprine and ovine. In this report, we characterize caprine and ovine RVAs based on full-length enterotoxin gene NSP4 (n=8) to identify their genotype difference and sequence variability.
Sampling, processing, and viral RNA extraction
The collection of samples was done from goat kids and lambs that were below six months of age, irrespective of signs of diarrhoea. Specimens were processed by preparing a 10% suspension in 1X PBS and stored further at -20°C for long time storage. The viral RNA was isolated using QIAzol Lysis Reagent (QIAGEN, USA) was suspended in 20 µl of nuclease-free water (NFW) (QIAGEN, Hilden, Germany), and kept at -20°C until further use.
cDNA preparation by Reverse Transcription
Initially, 15 µl (100-200 ng) of viral RNA was mixed with 1.0 µl (100 ng/µl) of random hexamer (QIAGEN, Hilden Germany) and 1 µl of DMSO (MB grade, Merck, Darmstadt, Germany) followed by incubation at 95°C for 5 min and 10 min snap chilling on ice. Simultaneously, 5X RT buffer, 1µl of 10mM dNTPs (QIAGEN, Hilden Germany), 1µl of (40U) RNase Inhibitor Ribolock™ (Fermentas, Vilnius, Lithuania), and 1µl of M-MLV RT enzyme (Promega, Madison, USA) were mixed accordingly and added to the snap chilled viral RNA mix making the final volume of the reaction to 25 µl and kept at 37°C for 60 min. The RT enzyme was inactivated by incubating the reaction at 80°C for 5 min. Thereafter, the cDNA prepared was employed in diagnosing RVA using custom-designed primers.
Molecular detection of rotavirus and NSP4 amplification
The RVA presence in the fecal samples was detected employing a degenerate primer pair based on the VP6 gene by the RT-PCR, as described earlier28. Consecutively, the positive samples were subjected to PCR amplification for the full-length NSP4 enterotoxin gene. We used published primers to amplify the full-length gene (742 bp) as described earlier 25. The primer name, sequence and the amplicon size of the VP6 and NSP4 gene used in the diagnosis and characterization of RVA isolates are given in Table 1. Following the amplification of desired products, the amplicons thus generated were cloned and sequenced in pDrive TA cloning vector according to a previous protocol29. The quality of each sequence thus generated was analyzed using the Finch TV (Geospiza, Inc. UK) software version 1.4, and the sequence identity was verified using the BLAST software in GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Table (1):
Primers used for the diagnosis and characterization of small ruminant origin RVAs using RT-PCR assay.
| Gene | Primer Name | Sequences 5’——- 3’ | Amplicon Size | References |
|---|---|---|---|---|
| VP6 | RVA-D-F | TTTGATCACTAAYTATTCACC | 226 bp | 28 |
| VP6 | RVA-D-R | GGTCACATCCTCTCACTA | ||
| NSP4 | NSP4 (1–28) [+] | GGCTTTWAAAAGTTCTGTTCCGAGAGAG | 743 bp | 25 |
| NSP4 | NSP4 (722–743) [-] | TAAGACCRTTCCYTCCATTAAC |
Eight isolates showing positive results in PCR were cloned and sequenced which was outsourced to M/s Eurofins Genomics, Bangalore. Out of the 08 isolates, four samples each from caprine and ovine origin were characterized. The sequence database generated on NSP4 genes was submitted to NCBI GenBank nucleotide repository under accession numbers MT998256 to MT998263. The strain name, place of collection, species, length, and accession numbers is given in (Table 2).
Table (2):
Randomly selected isolates from different states of Northern and Southern India were submitted in the NCBI GenBank database.
S. No. |
Strain Name |
Place of collection |
Species |
Length (bp) |
Accession No. |
|---|---|---|---|---|---|
1 |
K-UP86 |
Uttar Pradesh |
Caprine |
724 |
MT998256 |
2 |
K-UK95 |
Uttarakhand |
Caprine |
724 |
MT998257 |
3 |
K-KAR110 |
Karnataka |
Caprine |
714 |
MT998258 |
4 |
K-TN102 |
Tamil Nadu |
Caprine |
719 |
MT998259 |
5 |
L-TN76 |
Tamil Nadu |
Ovine |
647 |
MT998260 |
6 |
L-RAJ32 |
Rajasthan |
Ovine |
726 |
MT998261 |
7 |
L-RAJ54 |
Rajasthan |
Ovine |
750 |
MT998262 |
8 |
L-KAR65 |
Karanataka |
Ovine |
726 |
MT998263 |
Phylogenetic analysis
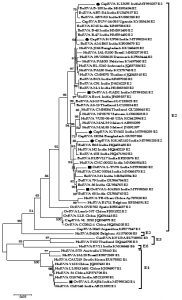
Phylogenetic analysis was performed for RVA NSP4 gene with different isolates from India and worldwide. A total of 08 strains of the current study were subjected to phylogenetic analyses with 57 sequences from different species all over the world retrieved from the NCBI database (Fig. 1). The analysis was done using the MEGA 7.0 software30. In both cases, the evolutionary history was obtained using the Maximum Likelihood (ML) method based on the Tamura-3 parameter model for computing the distances with the support of 1000 bootstrap replicates. A separate gamma distribution was used to model evolutionary frequency differences between the sites.
Per cent identity calculation and comparison of Inter-Species Variable Domain (ISVD) region
For per cent identity calculation, we analyzed current study sequences in MegAlign software packaged in DNASTAR where sequences were aligned by ClustalW, and individual distances among them were calculated. Furthermore, the rotavirus genotyping web-based tool hosted by ViPRBRC for RVA classification (https://www.viprbrc.org) was used to infer the E genotypes of all NSP4 sequences. The ORFs of all individual genes were translated theoretically using the EditSeq tool of DNASTAR. A comparison of the pre-defined interspecies variable domain (ISVD) region was made with the assistance of the Protean tool the DNASTAR software package.
Phylogenetic analysis
The phylogenetic analysis of 8 NSP4 genes of the current study sequence revealed 7 (4 caprine and 3 ovines) of E2 genotype and 1 (ovine) E1 genotype (Fig. 1). This observation was also confirmed by the web-based rotavirus genotyping tool of ViPRBC. As per the previous literature E1 genotype for ovine RVA strain has never been reported in India earlier. In phylogenetic analysis, all the E2 type strains of the current study clustered inside a major clade comprising different E2 genotype isolates from worldwide. Out of the four caprine strains of the current study, three-branched alongside bovine RVA isolates from India whereas one appeared alongside a Bangladeshi caprine RVA strain. Four ovine strains of the current study branched differently; two isolates from Rajasthan branched with bovine RVA isolates from India. At the same time, one each from Tamil Nadu and Rajasthan were closely related to human RVA isolates from India. The ovine RVA strain L-RAJ54 clustered inside the minor clade comprising human RVA of the E1 genotype.
Fig. 1: Phylogenetic analysis of small ruminant RVA based on the full-length NSP4 gene. Caprine and ovine strains have been depicted with a black circle (●) and black square (■) respectively. The tree is divided into E2 major and an E1 minor clade which comprises current study strains.
Percent identity analysis
The per cent identity analysis was done with the selected reference NSP4 genotypes which have been reported in humans and domesticated artiodactyl type species like bovine and porcine. The nucleotide per cent identity analysis revealed that all the E2 genotype strains of caprine and ovine species of the current study showed the highest (88.4% to 90.4%) and equal per cent identity with two reference human RVA isolates DS-1 (AF174305) and AU-1 (D89873) (Table 3). Ovine strain L-RAJ54 showed the highest nucleotide similarity percentage (91.3%) with reference human RVA E1 genotype isolate Wa (K02032). The current study caprine and ovine strains shared 87.4 to 93.8% and 77.7 to 95.1% of nucleotide similarity among themselves, respectively. This showed that ovine samples were more divergent among them with respect to caprine samples.
Table (3):
Nucleotide similarity index of current study RVA NSP4 strains with reference isolates.
CapRVA_K-UP86_E1 |
CapRVA_K-UK95_E1 |
CapRVA_K-KAR110_E1 |
CapRVA_K-TN102_E1 |
OvRVA_L-TN76_E1 |
OvRVA_L-RAJ32_E1 |
OvRVA_L-RAJ54_E2 |
OvRVA_L-KAR65_E1 |
Human_Wa_ 1974_E1 |
Human_DS-1_1976_E2 |
Human_AU-1_1982_E3 |
Human_B4106_ 2000_E5 |
HuRVA_RV11 _2011_E6 |
BoRVA_PP-1_2001_E8 |
PoRVA_CMP034 _E9 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CapRVA_K-UP86_E1 |
*** |
93.8 |
89.5 |
90.8 |
91.3 |
94.6 |
78.5 |
90.9 |
81.6 |
90.4 |
90.4 |
84.3 |
80.1 |
70.1 |
79.8 |
CapRVA_K-UK95_E1 |
*** |
87.4 |
87.9 |
89.2 |
92.8 |
77.6 |
88.5 |
80.5 |
88.9 |
88.9 |
82.6 |
79.1 |
70.3 |
78.7 |
|
CapRVA_K-KAR110_E1 |
*** |
91.9 |
92.7 |
89.4 |
76.8 |
93.4 |
79.6 |
88.6 |
88.6 |
82.2 |
78.2 |
70.3 |
77.9 |
||
CapRVA_K-TN102_E1 |
*** |
91.8 |
89.8 |
78.4 |
93.3 |
81.2 |
88.4 |
88.4 |
82.9 |
79.1 |
71.4 |
79 |
|||
OvRVA_L-TN76_E1 |
*** |
91.2 |
78.1 |
95.1 |
80.4 |
89.8 |
89.8 |
82.1 |
77.9 |
70.1 |
78.2 |
||||
OvRVA_L-RAJ32_E1 |
*** |
77.7 |
90.6 |
80.6 |
89.1 |
89.1 |
83.3 |
80.2 |
70.7 |
78.9 |
|||||
OvRVA_L-RAJ54_E2 |
*** |
78.5 |
91.3 |
79.5 |
79.5 |
80.8 |
79.6 |
71.4 |
83.2 |
||||||
OvRVA_L-KAR65_E1 |
*** |
81.1 |
89.8 |
89.8 |
83.9 |
79.6 |
71.6 |
79.2 |
|||||||
Human_Wa_1974_E1 |
*** |
82 |
82 |
82.7 |
82 |
76 |
86.5 |
||||||||
Human_DS-1_1976_E2 |
*** |
100 |
84.1 |
80.2 |
70.7 |
79.3 |
|||||||||
Human_AU-1_1982_E3 |
*** |
84.1 |
80.2 |
70.7 |
79.3 |
||||||||||
Human_B4106_2000_E5 |
*** |
80.7 |
70.9 |
81.6 |
|||||||||||
HuRVA_RV11_2011_E6 |
*** |
71 |
79.2 |
||||||||||||
BoRVA_PP-1_2001_E8 |
*** |
71.8 |
|||||||||||||
PoRVA_CMP034_E9 |
*** |
Amino acid sequence comparison of RVA NSP4 ISVD
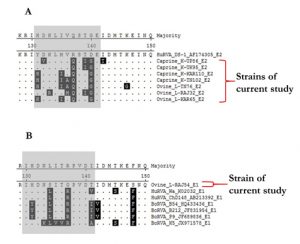
The NSP4 strains were translated theoretically and aligned using the MegAlign software to observe any crucial changes in the ISVD region of NSP4 C-terminal (Fig. 2). The observations revealed that E2 genotype strains of current study strains contain more variation in comparison to E1 type when aligned along with reference isolates. E2 type strains showed some number of varied amino acid residues in the ISVD region (131-141 aa) whereas the single E1 type ovine strain L-RAJ54 showed variance at residue position 131, 134, 137, and 141 only.
Fig. 2. Alignment of an interspecies variable domain (ISVD) of NSP4 from the current study and reference isolates. (A) Alignment depicts the amino acid sequence variation in E2 type strains (B) Alignment depicts the amino acid sequence variation in E1 type strains. Variable residues have been highlighted in black colour showing variance in sequences.
RVAs are known to cause diarrhoea in human neonates31, calves, and piglets32, but their aetiology has not been studied well in small ruminants. The current study reports the presence of RVAs in the caprine and ovine population of India along with the characterization of its major enterotoxin gene for the first time in India. However, few reports of prevalence emerged from India in the past for small ruminants33-37 but these reports failed to provide data on sequence-based characterization of RVA in small ruminants. Therefore, this study was designed to decipher the circulating potential of RVA in the small ruminant population of India as well as to characterize them based on sequence and phylogeny.
The phylogenetic analysis revealed a diverse population of ovine and caprine RVA based on the full-length NSP4 gene where in seven E2 type strains of the present study branched into different sub-clusters within the major E2 clade. One ovine RVA sample showed E1 genotype specificity which was found closer to a human RVA strain ChD148 (GenBank record only) of India. These observations point towards the diverse origin of RVAs in the Indian small ruminant population. Closeness to human and bovine type RVA isolates also suggest the ongoing interspecies transmission occurring between these ungulates and other mammals including humans. We aligned and observed the NSP4 amino acid sequences of the present study and found that strains possess a significant number of amino acid residue changes in the interspecies variable domain of the NSP4 C-terminus. This observation also supports the regarding the sequence and phylogenetic variation among small ruminant RVAs. Nevertheless, it will be interesting to observe whether these amino acid changes will lead to any alteration in the infectivity of these viral strains.
Overall the data presented in the study hypothesize that RVA strains circulating in these small ruminant populations may share a common origin which is usually found in artiodactyl species, but future studies are needed to confirm these findings. This hypothesis is reasonable because humans, large and small ruminants share a common dwelling place. The data from the prevalence also warrants strict surveillance measures to be taken in the future to know about the circulating genotypes of small ruminant RVAs.
ACKNOWLEDGMENTS
Authors acknowledge and express thanks to their respective Institutes and Universities. Help of Shanmuganathan in sampling is acknowledged. YSM acknowledges the Education Division, ICAR, Govt. of India for National Fellowship.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All the authors substantially contributed to the conception, compilation, checking, and approving the final version of the manuscript, and agree to be accountable for its contents.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files.
- Desselberger U. Rotaviruses. Virus Res. 2014;190:75-96.
Crossref - Dhama K, Chauhan R, Mahendran M, Malik S. Rotavirus diarrhea in bovines and other domestic animals. Vet Res Commun. 2009;33(1):1-23.
Crossref - Martella V, Decaro N, Buonavoglia C. Enteric viral infections in lambs or kids. Vet Microbiol. 2015;181(1-2):154-160.
Crossref - Papp H, Malik YS, Farkas SL, Jakab F, Martella V, Banyai K. Rotavirus strains in neglected animal species including lambs, goats and camelids. Virus Disease. 2014;25(2):215-222.
Crossref - Malik YS, Sircar S, Dhama K, et al. Molecular epidemiology and characterization of picobirnaviruses in small ruminant populations in India. Infect Genet Evol. 2018;63:39-42.
Crossref - Bachofen C, Vogt H-R, Stalder H, et al. Persistent infections after natural transmission of bovine viral diarrhoea virus from cattle to goats and among goats. Vet Res. 2013;44(1):32.
Crossref - Amer HM. Bovine-like coronaviruses in domestic and wild ruminants. Anim Health Res Rev. 2018;19(2):113-124.
Crossref - Ball JM, Tian P, Zeng CQ-Y, Morris AP, Estes MK. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272(5258):101-104.
Crossref - Zhang M, Zeng CQ-Y, Morris AP, Estes MK. A functional NSP4 enterotoxin peptide secreted from rotavirus-infected cells. J Virol. 2000;74(24):11663-11670.
Crossref - Dong Y, Zeng CQ-Y, Ball JM, Estes MK, Morris AP. The rotavirus enterotoxin NSP4 mobilizes intracellular calcium in human intestinal cells by stimulating phospholipase C-mediated inositol 1, 4, 5-trisphosphate production. Proceedings of the National Academy of Sciences. 1997;94(8):3960-3965.
Crossref - Hyser JM, Collinson-Pautz MR, Utama B, Estes MK. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. MBio. 2010;1(5).
Crossref - Tian P, Estes MK, Hu Y, Ball JM, Zeng C, Schilling WP. The rotavirus nonstructural glycoprotein NSP4 mobilizes Ca2+ from the endoplasmic reticulum. J Virol. 1995;69(9):5763-5772.
Crossref - Newton K, Meyer JC, Bellamy AR, Taylor JA. Rotavirus nonstructural glycoprotein NSP4 alters plasma membrane permeability in mammalian cells. J Virol. 1997;71(12):9458-9465.
Crossref - Au K-S, Mattion NM, Estes MK. A subviral particle binding domain on the rotavirus nonstructural glycoprotein NS28. Virology. 1993;194(2):665-673.
Crossref - O’brien JA, Taylor JA, Bellamy A. Probing the structure of rotavirus NSP4: a short sequence at the extreme C terminus mediates binding to the inner capsid particle. J Virol. 2000;74(11):5388-5394.
Crossref - Horie Y, Nakagomi O, Koshimura Y, et al. Diarrhea induction by rotavirus NSP4 in the homologous mouse model system. Virology. 1999;262(2):398-407.
Crossref - Jagannath M, Kesavulu M, Deepa R, et al. N-and C-terminal cooperation in rotavirus enterotoxin: novel mechanism of modulation of the properties of a multifunctional protein by a structurally and functionally overlapping conformational domain. J Virol. 2006;80(1):412-425.
Crossref - Matthijnssens J, Ciarlet M, McDonald SM, et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol. 2011;156(8):1397-1413.
Crossref - RCWG. Rotavirus Classification Working Group: Newly assigned genotypes. https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg
- Ciarlet M, Liprandi F, Conner M, Estes M. Species specificity and interspecies relatedness of NSP4 genetic groups by comparative NSP4 sequence analyses of animal rotaviruses. Arch Virol. 2000;145(2):371-383.
Crossref - Araujo IT, Heinemann MB, Mascarenhas JDAP, Assis RMS, Fialho AM, Leite JPG. Molecular analysis of the NSP4 and VP6 genes of rotavirus strains recovered from hospitalized children in Rio de Janeiro, Brazil. J Med Microbiol. 2007;56(6):854-859.
Crossref - Fredj MBH, Zeller M, Fodha I, et al. Molecular characterization of the NSP4 gene of human group A rotavirus strains circulating in Tunisia from 2006 to 2008. Infect Genet Evol. 2012;12(5):997-1004.
Crossref - Gonzalez-Ochoa G, Menchaca GE, Hernandez CE, Rodriguez C, Tamez RS, Contreras JF. Mutation distribution in the NSP4 protein in rotaviruses isolated from Mexican children with moderate to severe gastroenteritis. Viruses. 2013;5(3):792-805.
Crossref - Tavares TdM, Brito WMEDd, Fiaccadori FS, et al. Molecular characterization of the NSP4 gene of human group A rotavirus samples from the West Central region of Brazil. Mem Inst Oswaldo Cruz. 2008;103(3):288-294.
Crossref - Malik YS, Kumar N, Sharma K, et al. Molecular analysis of non structural rotavirus group A enterotoxin gene of bovine origin from India. Infect Genet Evol. 2014;25:20-27.
Crossref - Saurabh S, Sircar S, Kattoor JJ, et al. Analysis of structure-function relationship in porcine rotavirus A enterotoxin gene. J Vet Sci. 2018;19(1):35-43.
Crossref - Mori Y, Borgan MA, Ito N, Sugiyama M, Minamoto N. Sequential analysis of nonstructural protein NSP4s derived from group A avian rotaviruses. Virus Res. 2002;89(1):145-151.
Crossref - Mondal A, Sharma K, Malik YS, Joardar SN. Detection of group a rotavirus in faeces of diarrhoeic bovine porcine and human population from eastern India by reverse transcriptase-polymerase chain reaction. Adv Anim Vet Sci. 2013;1(1S):18-19.
- Kattoor JJ, Saurabh S, Malik YS, et al. Unexpected detection of porcine rotavirus C strains carrying human origin VP6 gene. Vet Q. 2017;37(1):252-261.
Crossref - Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870-1874.
Crossref - Estes MK. Rotaviruses. Fields Virology. 2007:1917-1974.
- Papp H, Laszlo B, Jakab F, et al. Review of group A rotavirus strains reported in swine and cattle. Vet Microbiol. 2013;165(3-4):190-199.
Crossref - Singh U, Singh R, Singh AP, Yadav SK, Sircar S, Malik YS. Detection and characterization of caprine and ovine rotaviruses, India. Indian J Anim Sci. 2017;87(11):1358-1361.
- Reddy G, Kumari A, Mishra A, et al. Prevalence of groupa rotavirus in diarrhoeic goat kids from organized goat farms. Indian J Comp Immunol Microbiol Infect Dis. 2014;35(1):9-12.
- Wani S, Bhat M, Nawchoo R, Munshi Z, Bach A. Evidence of rotavirus associated with neonatal lamb diarrhoea in India. Trop Anim Health Prod. 2004;36(1):27-32.
Crossref - Gazal S, Taku A, Kumar B. Predominance of rotavirus genotype G6P [11] in diarrhoeic lambs. Vet J. 2012;193(1):299-300.
Crossref - Gupta V, Pandey M, Nayak A, Rajoriya S, Bordoloi S. Polyacrylamide gel electrophoresis and silver staining for detection of rotavirus in goat fecal samples. Journal of Pharmacognosy and Phytochemistry. 2019;8(3):472-475.
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.