ISSN: 0973-7510
E-ISSN: 2581-690X
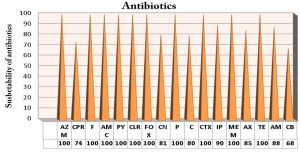
Typhoid fever is a paramount reason for horribleness that more mortal sin “around the sum ages aggregations clinched alongside iraq it initiated by salmonella typhi. Salmonella typhi is diagnosed serologically by the Widal test and confirmed by vitek and using polymerase chain reaction (PCR) based amplification of DNA from the bacterial samples of typhoid fever patients. The present study was designed to detect class I integron gene encoding antimicrobial of S. typhi using appropriate primers by PCR. These isolates of this study were collected from postgraduate laboratories (Prepared samples in vitro prepared diagnostics), they were a previous collected from carried out in Al Najaf provenance, throughout those period from July 2018 on March 2019 including 231 cases from blood, stool samples collected from patients suffering from typhoid fever were attended to Al-Sader Medical City and Al-Hakim General Hospital in Al-Najaf province. Biochemically tests and monovalent antisera gave 117 (50.64%) positive result S. typhi isolates and confirmed by Vitek system and PCR which showed positive result 59 (50.42%). Fifty nine isolates of S. typhi, were collected from patients with typhoid fever that distributed to 40/59 (34 %) from blood , 19/59 (15.1%) stool. Molecular detection revealed that most isolates of S. typhi were positive results to (intI) gene 43/59 isolate (the specific primer (intI) gene for S. typhi bacteria was designed in this study by using bioinformatics programs with NCBI website). According to the different diagnostic above, Vitek and PCR method were more sensitivity technique for S. typhi detection among typhoid patients. The results of virulence factors of S.typhi isolates were negative results for gelatinase, hemolysin, protease and capsulated. Multidrug resistance (MDR) of S. typhi isolates were represented by 18 antibiotics resistance to class and sub class of antibiotic. All S. typhi isolates appeared high resistance 100% to Aztreonam (AZM15), Nitrofurantion (F), Amoxicillin/clavulanicacid (AMC30), (PY25), Clarithromycin (CLR), Cefoxitin (FOX30), Penecillin(P10), Cefotaxime (CTX30), ampicillin (AMP), Meropenem (MEM), Tetracycline(TE30). Also resistance of isolates that revealed 91% to Impinem (IP ), 88% Ampicillin (AM10), 85%Amoxillin (AX), 81% Gentamicin (CN10), 80% Chloramphenicol (C30), 74% Cefpirome (CPR) and 68% Carbenicillin (CB).
Salmonella typhi, Typhoid fever, PCR, integron class I, intI gene, Virulence Factors
Typhoid fever is an acute illness, potentially fatal systemic caused by Salmonella typhi and Salmonella paratyphi, pathogen only specific to humans. It has been shown that typhoid occurs in many developing countries, which it still remains an important cause of mortality and morbidity1. Typhoid fever is still a major health concern in the developing world as it is found endemically throughout Asia Africa, particularly in South Asia,East Asia and South America. In a broader sense, typhoid fever is categorized under enteric fever which is described as a systemic illness described by abdominal pain, fever and other symptoms as vomiting, nausea, anorexia and headache2. When enteric fever is caused by Salmonella typhi, it is known as typhoid fever and when it is due to Salmonella Paratyphi A, B, or C, it is called paratyphoid fever.
Salmonella typhi bacterium are an obligate parasite with no other known reservoir outside of humans. A 100,000 organisms of Salmonella typhi make up an infectious dose, and the disease is typically spread through feces and urine of infected people in contaminated food and water2. Typhoid is transmitted by the fecal-oral route, in most cases an asymptomatic carrier of S.typhi, or an individual who has recently recovered from the infection, continues to excrete large number of organisms in the stool and contaminates food or water, either through direct food handling, through transfer of bacteria by flies and other insects, or by contamination of poTable water3. Approximately 10% of patients recovering from typhoid fever excrete S.typhi in the stool for three months, and in the 2-3% became permanent carriers4.
Salmonella typhi contains an endotoxin typical of gram negative organisms, as well as the Vi antigen which is thought to increase virulence. Many of the S.typhi virulence factors, such as toxin genes invasion and adhesion are clustered in certain areas of the chromosome known as Salmonella pathogenicity islands5. Virulence factors present on the surface of Salmonella typhi such as lipopolysaccharides, capsule and fimbriae can affect the virulence of the bacterium, and also are key targets of the host immune system6. Endotoxin play an important role in the pathogenesis of Salmonella typhi infection during the bacteremia stages of typhoid fever that produced as a result of the acting endotoxin directly and indirectly through the release of endogenous pyrogens from leukocytes7.
Most Salmonella strains carry a large number of plasmids that contains virulence genes that are essential to trigger systemic disease, their involvement in the enteric stage of the infection is unknown8. Antimicrobials is important for the control and treatment of Salmonella spp. However, antimicrobial- and multidrug-resistant Salmonella strains have emerged, leading to treatment failure9. Many Strains of Salmonella typhi resistant to all three first-line drugs were in existence,with the development of quinolone resistance, third-generation cephalosporin’s were used for treatment, but sporadic reports of resistance to them also followed10,11.
Multidrug resistance is an increasing threat to global health. There are > 14 million cases of typhoid fever every year and > 135,000 deaths, the disease is primarily controlled by antimicrobial treatment, but this is becoming increasingly difficult due to MDR11.Multidrug-resistant typhoid fever is defined as typhoid fever caused by Salmonella typhi are resistant to all the three first-line recommended drugs for treatment, including cotrimoxazole ampicillin and chloramphenicol12. ESBL-producing Salmonella isolates resistant to several antibiotic agents,third-generation cephalosporin’s referred to extended spectrum cephalosporin’s were effective against ampicillin hydrolyzing β-lactamases and gained widespread use in clinical use. They include ceftriaxone, ceftazidime, cefotaxime and others. Treatment of Salmonella typhi with third-generation cephalosporin, azithromycin and fluoroquinolones drugs are the main treatment13.
Integron are genetic elements that contain the genetic determinant for the components of asite-specific recombination system that recognizes and captures mobile gene cassettes. An integron is a gene capture system found on plasmids, on chromosome and in transposon, platforms that incorporate exogenous open reading frames by site-specific recombination and convert them to functional genes by ensuring their correct expression14. Integron can be divided into two major group : resistance integrons and super-integrons Although not independently mobile, integrons are widespread versatile DNA elements and can be divided into two distinct subsets: the mobile integrons and the chromosomal integron15.
Integrons are classified based on sequence similarity, there are at least five classes of integrons with class I integrons being the most studied and characterized 16. Class 1 integrons are the most common and widely distributed among Gram-negative bacteria and are associated with functional and nonfunctional transposons17,18. Class 1 integron is composed of the(3‟CS) and including 2 (ORFs)open reading frames. QacE1 gene, it confers resist to qac quaternary ammonium compound; and Soul gene is confer resist to sulphonamides. There are responsible for the mobility of transposons and also found in Salmonella typhi19. PCR is rapid and sensitive assay used to detect microbial pathogens in clinical samples. PCR method had diagnostics value for detection of Salmonella among all the diagnostic tests20-22. The present study carried out to achieve the following objects: 1- Isolation and identification of Salmonella typhi collected from patients with typhoid fever from blood and stool by Vetik and PCR diagnosis . 2- Determenation of the virulence factors gelatin and protease among clinical isolates. 3- determination of MDR of the antimicrobial susceptibility in S.typhi isolates. 4- Detection of the Class I integron (intI) gene in S.typhi isolates.
Samples collection
These isolates of this study were collected from postgraduate laboratories(Prepared samples in vitro prepared diagnostics), they were a previous collected from 231 samples were involved 122 stool samples and 109 blood samples. Those period from July 2018 on March 2019 were attended to Al-Hakim General Hospital and Al-Sader Medical City Hospital in Al-Najaf province. It collected from patients were suffering from typhoid fever and swabbed onto Xylose-Lysine-Desoxycholate Agar (XLD) and MacConkey agar media were incubated overnight.
Identification of Salmonella typhi isolates
- Microscopic Properties: Gram’s stain was used to examine the isolated bacteria for studying the microscopic properties as initial identification of S.typhi.23
- Cultural Characteristics: Morphological characteristics of Salmonella typhi were recorded on the specific media for primary identification23.
- Biochemical Tests: 1-2 colony of S.typhi isolates were tested for oxidase, Simmone Citrate, catalase, Kligler Iron agar and Indole tests.
- VITEK@2 GN ID Card: Salmonella typhi isolates were confirmed with the automated VITEK@2 compact system by using GN ID cards. It was carried out on each bacterial isolate to complete the final identification. The GN ID card is based on established biochemical (64 reaction) methods and newly developed substrates, measuring various metabolic activities (BioMerieux Company/http://www.bioMérieux.com).
C – Molecular Profiling of Class I Integron intI gene in Salmonella typhi isolates
- Bioinformatics of intI gene: Bio-statistical service technology and Bioinformatics has advance of diagnostics and medical in (NGS) and ecological research. Bioinformatics specialized in Microbiology and set new standards for high level services of data analysis including high expertise in NGS18,24. In this study is using Bioinformatics program to designing the specific primer of Class I intI gene in MDR Salmonella typhi.
- Genomics Bacterial DNA Extraction: DNA (Promega /USA) Extraction of Bacteria DNA is (59) of Salmonella typhi bacterium. The wizard chromosomal DNA purification kit is designed for extraction of Salmonella DNA.
- Polymerase Chain Reaction: PCR has been amplifying specific primer of Class I integron intI gene with genomic DNA of all isolates S.typhi. DNA purification kit was designed for extraction of S.typhi DNA. Gel-electrophoresis was used for detection of DNA by UV transilluminator25. The PCR assay was detected the (intI) gene. These specific primer synthesized by Alpha DNA company, Canada. the DNA marker and PCR product were resolved (1.5%) agarose gel by electrophoresis.
- Multidrug Resistance Tests in Salmonella typhi isolates: Antimicrobial susceptibility tests assay, as described by CLSI26. It was used antibiotics disks assay and Muller Hinton Agar medium in current study to the susceptibility of 18 antibiotics commonly including as Aztreonam(AZM15), Nitrofurantion(F), Amoxicillin/clavulanicacid (AMC30), (PY25), Clarithromycin(CLR), Cefoxitin(FOX30), Penecillin(P10), Cefotaxime(CTX30), ampicillin(AMP), Meropenem(MEM), Tetracycline(TE30), Impinem(IP), Ampicillin(AM10), Amoxillin(AX), Gentamicin (CN10), Chloramphenicol(C30), Cefpirome(CPR) and Carbenicillin(CB)26.
Virulence factors of Salmonella typhi
- Detection of Hemolysin: This agar medium was inoculated with S.typhi isolates, and incubated for 24/48 hr, at 37˚C. Positive results that show the clear zone around the bacterial colony appears which is referred to β –hemolytic and α- hemolytic or green zone referred to but negative results weren’t that show 27.
- Detection of Protease Enzyme: S.typhi isolates were inoculated the Skim agar medium, it was incubated for 24-48hr,at 37˚C. the positive result it was observed when clear zone was appeared around the colony while the negative results weren’t that show28.
- Detection of gelatin production: The specific media for detection gelatinase liquefaction was inoculated with a single colony of overnight culture from nutrient medium and incubated at 37 C°, for 24 hr of incubation period by placing the tube in refrigerator for (30)min. A positive result was indicates if the media was liquid which refer to ability of bacteria to produced gelatinase and negative results was solidify because of no ability to produced it29.
Statistical Analysis
In the current study using the program of SPSS computing (version 24) for the data analysis.
Identification of Salmonella typhi Isolates
During the current study period were collected 231 clinical samples from (122) blood samples and, (109) stool samples from patients with typhoid disease. Identification of Salmonella typhi isolates have been depending on initial identification the morphological, biochemical tests, and microscopically. Microscopically identification of S.typhi was appeared gram negative bacilli singly or in pairs, peritrichous flagellated, motile, non-spore forming bacteria, and non-capsulated. While the Morphological of S.typhi isolates were used the specific media such as XLD agar and MacConkey agar, appeared on culture medium once appeared the typical characteristics at 37°C after 18-24h, on blood agar S.typhi colonies appeared smooth, rounded,convex, and nonhemolytic and grey white color. But on MacConkey agar were appeared pale yellow ( non-lactose ferment ), 1-3mm in diameter and after 18-24 hour at 37°C. as well as, good growth of S.typhi colonies on XLD agar were appeared gray color with black center colonies because of it was ability to produce the H2S, as showed in [Fig. 1].
Biochemically tests results of S.typhi isolates were revealed on TSI, Sugars, Oxidase, Indole, Ureases and simmone citrate test. In TSI slants test, the slant and butt turned into AKL/ACID red and yellow colour respectively indicating to non-fermentation of glucose on slant and production of acid with H2S in the bottom. Glucose, mannitol, L-arabinose and sorbitol sugars were fermented by all isolates. Other tests of S.typhi isolates were appeared negative result for oxidase test, indole production, urease production and citrate utilization as showed in Table [Table.1].
Table (1):
Biochemical tests of Salmonella typhi isolates.
NO |
Biochemical tests |
Salmonella typhi results |
|---|---|---|
1. |
TSI test |
AKL/ACID with H2S |
2. |
Glucose, mannitol, L-arabinose sorbitol |
+ Sugars fermented |
3. |
Oxidase test |
– |
4. |
Indole test |
– |
5. |
Ureases test |
– |
6. |
simmone citrate slant test |
– |
7. |
Motility |
Motile + |
8. |
Spore |
– |
9. |
H2S Production |
+ |
VITEK@2GNID Cards System results of S.typhi isolates, the identification were included many biochemical tests. The results revealed S.typhi were revealed with cards ID the ranging excellent of isolates (percentage% from 95 to 99%). The 59 Salmonella typhi colonies on MacConkey agar.
Detection virulence factors of Salmonella typhi
The ability of (59) S. typhi to produce many virulence factors. All isolates of S.typhi have been produce the non-hemolytic on blood medium and non-capsulated. Likewise S.typhi isolates have no ability to produce of Gelatinase on specific gelatin media as well as it has non-produced of Proteinase on Skim agar medium.
Multi-drug Resistance of Salmonella typhi isolates
Among 59 Salmonella typhi isolates, they were susceptible to describe this study by susceptibility of 18 antibiotics, including as high resistance of Salmonella typhi isolates appeared high resistance 100% to Aztreonam (AZM15), Nitrofurantion (F), Amoxicillin/clavulanicacid (AMC30), (PY25), Clarithromycin (CLR), Cefoxitin (FOX30), Penecillin (P10), Cefotaxime (CTX30), ampicillin (AMP), Meropenem (MEM), Tetracycline(TE30). Also resistance of isolates that revealed 91% to Impinem (IP ), 88% Ampicillin (AM10), 85%Amoxillin (AX), 81% Gentamicin (CN10), 80% Chloramphenicol (C30), 74% Cefpirome (CPR) and 68% Carbenicillin (CB).as mentioned in [Fig. 2]. They have been need an overall increment in MDR typhoid created by S.typhi on different antimicrobial operators. A high variables would responsible, those development MDR isolates encountered with urban decay because of deindustrialization, government lodgin and engineering imagined.
Molecular Characteristics Salmonella typhi isolates
Characteristics of Class I Integron intI Gene of S.typhi isolates
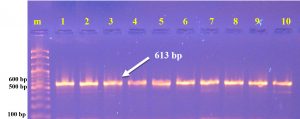
The present investigation indicates that the vicinity about a integron might have been altogether connected with 43(73 %) MDR isolates. Those distinguished genes encodes (integrase) from claiming class 1 integron. Specific primer designed in this study within NCBI web/ Bioinformatics program. PCR was used to dtected (intI) gene of Salmonella typhi. The results of Salmonella typhi diagnosis by PCR technique that 43/59(73%) isolates visualized in [Table 2] and [Fig. 3].
Table (2):
The Sequence and concentration of intI primers.
No |
Primers |
Primers sequences 3- 5 |
Product size |
Temperature Annealing |
References |
|---|---|---|---|---|---|
1. |
intI- F intI- R |
ACCCCCGTAATGGCTAACTT GCAGTTGCAAACCCTCACTG |
613 bp |
60°C |
Primer designed in this study |
The current study identified of Salmonella typhi isolates were depended on, microscopically, the morphology, biochemical tests and initial identification. Salmonella typhi colonies on XLD agar were appeared gray color with black center colonies. Results from previous studies suggested that iNTS disease and Typhoid fever were major causes of invasive bacterial febrile illness in the sampled locations, most commonly aff ecting child in both high and low population density30.Typhoid fever infection was caused by S.typhi and was transmitted through the oral route by the contaminated poultry, meat,, milk and eggs. Disease is most common in young child and elderly with peak incidence in summer and fall rainy season31.
Biochemically tests results of Salmonella typhi were observed negative result to simmons citrate, indole production, urease oxidase and H2S-production and TSI was ALK/ACID for all isolates.It was founded S. typhi isolates couldn’t produce Urease enzyme. All biochemically result were agreed with32-34.
All results of virulence factors of Salmonella typhi showed that all isolates didn’t produced genelatinase, hemolytics and proteinase enzymes. Thus, entero-chelin secretion in vivo could be a vital step in the stages of the infection cycle in which intestine – population, penetrate, the mucous layer covered the intestinal, translocate out of intestinal lumen through the epithelial cells, and finally step spread to organs and they may survive35.
MDR results in this study by susceptibility of (18) antibiotics, including as high resistance of Salmonella typhi isolates 100% to Aztreonam(AZM15), Nitrofurantion(F), Amoxicillin/clavulanicacid (AMC30), (PY25), Clarithromycin (CLR), Cefoxitin(FOX30), Penecillin (P10), Cefotaxime (CTX30), ampicillin (AMP), Meropenem (MEM), Tetracycline (TE30). Antibiotics resistance in S.typhi is a threat to human public health36. In previously studies, MDRSalmonella bacteria were frequently observed among pigs,chickens and ducks. In addition, MDR Salmonella was serotype dependent37. As well as Salmonella enteritidis and Salmonella typhimurium were strongly association with MDR antibiotics phenotypes. MDR isolates could transfer to humans via animal or animal production and a great risk to public health38.
While, these isolates gave high resistance to other antimicrobial such as 91% to Impinem(IP), 88% Ampicillin(AM10), 85%Amoxillin(AX), 81% Gentamicin (CN10), 80% Chloramphenicol(C30), 74% Cefpirome(CPR) and 68% Carbenicillin(CB).as mentioned in [Fig.2]. Other studies mentioned that the 41 Salmonella typhi screened for antibiotics susceptibility, 25 (61.0%) were resistant to five or more antibiotics. Also, 84% of bacteria were isolated from patients who didn’t respond to initial treatments. The high percentage of MDRSalmonella typhi revealed was worrying and quite revealing39. MDR Salmonella typhi in Nigeria, Lagos. Periodic review of antimicrobial used in hospitals was essential as the efficacies of cotrimoxazole chloramphenicol and ampicillin were now doubtful. ofloxacin and Ciprofloxacin were effective antibiotics for treatment of typhoid infection, but with current trends of drug in Nigeria, resistance was likely to development. Therefore, provide an early warning signal for the prudent use of fluoroquinolone DRUGS to preserve their usefulness39.
Our results of molecular profiling of Class I Integron gene in S.typhi isolates, the present investigation indicates that the vicinity about a integron might have been altogether connected with 43/59 (73%) MDR isolates. Those distinguished genes encodes (integrase) from claiming class 1 integron. Integron were universal Around medication of Gram-negative clinical segregates. In addition, A percentage combinations for gene cassettes appear to be should need a nearby distribution, inasmuch as others appear will have a broad appropriation. Class 1 integron need aid spotted every now and again on plasmids that could make exchanged by conjugation. Those integron-carrying segregates would generally multidrug resistant, while those integron-lacking detaches would additional defenseless with drugs, Anyhow Likewise over other cases, not the sum safety determinant were spotted on Integrons40.
The conclusion of this study included into 1- Salmonella typhi isolates have been a big role in causing typhoid disease among clinical isolates. 2- Virulence factors of Salmonella typhi isolates have no ability to produce the gelatinase, proteinase and non-hemolytic among clinical isolates. 3- Genotypic detection by PCR revealed the presence of Class1 integron gene that have are relationships with MDR. 4- the current study revealed high resistance of S.typhi isolates have a reach to resistance 100% to Aztreonam (AZM15), Nitrofurantion (F), Amoxicillin/clavulanicacid (AMC30), (PY25), Clarithromycin (CLR), Cefoxitin(FOX30), Penecillin(P10), Cefotaxime (CTX30), ampicillin (AMP), Meropenem (MEM), Tetracycline (TE30).
ACKNOWLEDGMENTS
We would like to thank to Lab. Molecular Genetics of department of Medical Laboratory Techniques/AL-Toosi University college and department of Ecology in college of science / University of Kufa.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All data were analyzed during this work are included in the manuscript.
- Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Org. 2004;82(5):346-53.
- Capoor MR, Nair D. Quinolone and Cephalosporin Resistance in Enteric Fever. Journal of Global Infectious Diseases. 2010;2(3):258-262.
Crossref - Butler T. Treatment of Typhoid Fever in the 21st Century: Promises and Shortcomings. Clin Microbiol Infect. 2011;17(7):959-963.
Crossref - Zige DV, Ohimain EI, Sridhar MK. Community-based screening of asymptomatic typhoid carriers in Wilberforce Island, Bayelsa State, Nigeria. Int J Health Sci Res. 2013;3:119-26.
- Kaur J, Jain SK. Role of Antigens and Virulence Factors of Salmonella Enterica Serovar Typhi in Its Pathogenesis. Microbiol Research.2012;20;167(4):199-210.
Crossref - Robert W Crawford. Very Long O-Antigen Chains Enhance Fitness During Salmonella-Induced Colitis by Increasing Bile Resistance. Usaplos Pathog. 2012;8:E1002918.
Crossref - Luby SP. Bacteria: Salmonella Typhi and Salmonella Paratyphi. Encyclopedia of Food Safety. 2014;515-522.
Crossref - Kalule KJ, Kaddu-Mulindwa DH, Asiimwe BB. Antimicrobial Drug Resistance and Plasmid Profiles of Salmonella Isolates from Humans and Foods of Animal Origin in Uganda. Advances in Infectious Diseases. 2012;2(4):151-155
Crossref - Gong J, Xu M, Zhu C. et al. Antimicrobial resistance, presence of integrons and biofilm formation of Salmonella pullorum isolates from eastern China (1962-2010). Avian Pathol. 2013;42(3):290-294.
Crossref - Kumar R, Gupta N, Shalini. Multidrug Resistant Typhoid Fever. Indian J Pediatr. 2007;74:39-42.
Crossref - Browne AJ, Hamadani BHK, Kumaran EAP, et al. Drug-resistant enteric fever worldwide, 1990 to 2018: a systematic review and meta-analysis. BMC Medicine. 2020;18(1):1-22.
Crossref - Zaki SA, Karande S. Multidrug-resistant typhoid fever: a review,” J Infect Dev Ctries. 2011;5(5):324-337.
Crossref - Keddy KH, Sooka A, Letsoalo ME, et al. Sensitivity and specificity of typhoid fever rapid antibody tests for laboratory diagnosis at two sub-Saharan African sites. Bull. World Health Organ. 2011;8:640-647.
Crossref - Hall RM, Stokes HW. Integrons: Novel DNA Elements Which Capture Genes by Site-Specific Recombination. Genetica. 1993;90:115-132.
Crossref - Mazel D. Integrons: Agents of Bacterial Evolution. Nat Rev Microbiol. 2006;4(8):608-620.
Crossref - Ravi A, Avershina E, Ludvigsen J, L”Abee-Lund TM, Rudi K. Integrons in the Intestinal Microbiota As Reservoirs for Transmission of Antibiotic Resistance Genes. Pathogens. 2014;3(2):238-248.
Crossref - Koczura R, Mokracka J, Barczak A, Krysiak N, Kaznowski A. Association Between the Presence of Class 1 Integrons, Virulence Genes, and Phylogenetic Groups of Escherichia Coli Isolates from River Water. Microbial Ecology. 2013;65(1):84-90.
Crossref - Al-Fatlawy HNK, Aldahhan HA, Alsaadi AH. Phylogenetic of ERIC-DNA Fingerprinting and New Sequencing of Aeromonas Species and V. Cholerae DNA. Am J Appl Sci. 2017;14(10):955-964.
Crossref - Saenz Y, Vinue L, Ruiz E, Somalo S, Martinez S, Rojo-Bezares B. Class 1 Integrons Lacking QacED1 and Sul1 Genes in Escherichia Coli Isolates of Food, Animal and Human Origins. Vet Microbiol. 2010;144(3-4):493-497.
Crossref - Prakash P, Mishra OP, Singh AK, Gulati AK, Nath G. Evaluation of Nested PCR in Diagnosis of Typhoid Fever. J Clin. Microbiol. 2005;43(1):431-432.
Crossref - Ambati SR, Nath G, Das BK. Diagnosis of typhoid fever by polymerase chain reaction. Indian J Pediatr. 2007;74:909-913.
Crossref - Resendiz-Nava CN, Esquivel-Hernandez Y, Alcaraz-Gonzalez A, Castaneda-Serrano P, Nava GM. PCR Assays Based on invA Gene Amplification are not Reliable for Salmonella Detection. Jundishapur J Microbiol. 2019;12(2):e68764.
Crossref - Jawetz E, Melnick JI, Adelberg EA. Medical Microbiology, 28th ED. Appleton and Lange, USA. 2019.
- Ghatak S, Blom J, Das S, et al. Pan-genome analysis of Aeromonas hydrophila, Aeromonas veronii and Aeromonas caviae indicates phylogenomic diversity and greater pathogenic potential for Aeromonas hydrophila. Antonie Van Leeuwenhoek. 2016;109:945-956.
Crossref - Sambrook J, Russell RW. Molecular Cloning: A Laboratory Manual. 3rd Edn. Cold Spring Harbor Laboratory Press, N.Y. 2013.
- CLSI, Clinical and Laboratory Standards Institute. the Clinical and Laboratory Standards Institute antimicrobial susceptibility testing standards M02, M07, and M11. Informational Supplement. CLSI document Melvin P. Weinstein, MD. M100-Ed30E. Performance Standards for Antimicrobial Susceptibility Testing, 30th Edition. 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087 USA. 2020.
- Baron EJ, Peterson lR, Finegold SM. Baily and Scott’s. Diagnostic Microbiology. 9th ed. Company. USA. 1994.
- Elsner H-A, Sobottka I, Mack D, Claussen M, Laufs R, Wirth R. Virulence factors of Enterococcus faecalis and Enterococcus faecium blood culture isolates. Eur J Clin Microbiol Infect Dis. 2000;19:39-42.
Crossref - Collee JG, Fraser AG, Marmino BP, Simons A. Mackin and McCartney Practical Medical Microbiology. 14th ed. The Churchill Livingstone, Inc. USA. 1996.
- Marks F, Kalckreuth VV, Aaby P, et al. Incidence of invasive salmonella disease in sub-Saharan Africa: a multicentre population-based surveillance study. International Vaccine Institute, SNU Research Park, Seoul, South Korea. Lancet Glob Health. 2017;5: e310-23-e236.
- Jerrold R, Turner S. The gastrointestinal tract: Salmonellosis, in Kumar, V, Abbas, AK, Fausto, and Aster, JC, editors, Robins and Cotran Pathologic Basis of Disease, 8th edition India. Elsevier. 2010: 801.
- Collee JG, Marr W. Culture of bacteria, in Mackie and Cartney, Practical Medical Microbiology, Collee JG, Fraser, AG, Marmion, BP and Simmons, A editors, 14th edition,Curchill Livingstone, India. 2006;121-124.
- MacFaddin JF. Biochemical test for identification of medical bacteria, 3rd ed. William & Wilkins Co. London. 2000.
- Nalbantsoy A. Prevention and identification of salmonella. 2012;4:1614.1622.
- Khaled E, Elgayar MA, Kholy EI, Yasser M, Elmonem A, Shady M. Application of Salmonella typhi’s outer membrane (omp) in diagnosis of typhoid. Int J Curr Res. 2013:2037-2042
- Zhao X, Yang J, Zhang B, Sun S, Chang W. Characterization of Integrons and Resistance Genes in Salmonella Isolates from Farm Animals in Shandong Province, China. Front Microbiol. 2017;8:1300.
Crossref - Clemente L, Correia I, Themudo P, Neto I, Canica M, Bernardo F. Antimicrobial susceptibility of Salmonella enterica isolates from healthy breeder and broiler flocks in Portugal. Vet J. 2014(2);200:276-281.
Crossref - Ziech RE, Lampugnani C, Perin AP, et al. Multidrug resistance and ESBL-producing Salmonella spp. Isolated from broiler processing plants. Braz J Microbiol. 2016;47(1):191-195.
Crossref - Akinyemi KO, Smith SI, Bola-Oyefolu AO, Coker AO. Multidrug resistance in Salmonella enterica serovar typhi isolated from patients with typhoid fever complications in Lagos, Nigeria. Journal The Royal Institute of Public Health. Elsevier Ltd. Public Health. 2005;119(4):321-327.
Crossref - Fluit AC, Schmitz FJ. Resistance Integrons and Super- Integrons. Clin Microbiol Infect. 2004;10(4):272-288.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.