ISSN: 0973-7510
E-ISSN: 2581-690X
The emergence of multidrug-resistant hypervirulent Klebsiella pneumoniae (MDR-hvKp) strains has become a significant concern in healthcare settings worldwide. This study aims to elucidate the current landscape of MDR-hvKp infections in diabetic patients, shedding light on the challenges posed by these pathogens and highlighting the urgent need for concerted efforts in surveillance, prevention, and treatment to mitigate their impact on public health. This is the prospective study conducted over a period of 12 months. This study consisted all non-duplicate n = 500 different clinical samples from diabetic patients which were received for bacterial culture in the microbiology department during the study period. Determination of antimicrobial susceptibility and drug resistance was performed by conventional and molecular methods. Among Klebsiella pneumoniae Extended Spectrum Beta-Lactamase (ESBL) positive isolates of K. pneumoniae, 53 isolates showed presence of blaSHV (n = 53, 77.9%), blaTEM (n = 51, 75%) and blaCTX-M (n = 42, 61.7%), blaTEM with blaSHV positive for 31 isolates, blaTEM with blaCTX-M positive for 27 isolates and 19 isolates were positive for blaTEM with blaSHV and blaCTX-M. Among 32 Metallo-β-lactamase (MBL) positive K. pneumoniae, blaKPC was positive for (n = 32, 47%), blaVIM + blaIMP (n = 31, 45.5%), blaVIM (n = 28, 41.1%), blaIMP (n = 24, 35.2%) and blaKPC + blaVIM (n = 23, 33.8%) were identified. The increasing prevalence of antibiotic resistance is limiting the potential treatment choices for diseases caused by bacteria that have developed resistance to drugs.
Klebsiella pneumoniae, Multidrug-resistant, ESBL, MBL, Hyper-virulent
The emergence of multidrug-resistant hypervirulent Klebsiella pneumoniae (MDR-hvKp) strains has become a significant concern in healthcare settings worldwide.1 Klebsiella pneumonia (K. pneumoniae), a common Gram-negative bacterium, has long been associated with various infections, ranging from urinary tract infections to severe pneumonia.2
K. pneumoniae represents a critical challenge in healthcare, particularly among diabetic patients. Diabetic patients, already predisposed to infections due to their compromised immune status and impaired wound healing, are particularly vulnerable to the deleterious effects of MDR-hvKp infections. The intertwining of multidrug-resistance and hypervirulence in these strains not only amplifies the difficulty in treating infections but also exacerbates the potential for rapid disease progression and mortality in diabetic individuals.3
Among the diverse patient populations affected by MDR-hvKp, individuals with diabetes mellitus are particularly vulnerable. Diabetes mellitus, characterized by hyperglycemia and impaired immune function, creates a conducive environment for bacterial infections, making diabetic patients prone to a higher risk of acquiring and experiencing severe complications from MDR-hvKp infections.4
Understanding the epidemiology, clinical manifestations, and mechanisms underlying the emergence and dissemination of MDR-hvKp in diabetic patients is imperative for guiding effective therapeutic strategies and infection control strategies.5
This study aims to elucidate the current landscape of MDR-hvKp infections in diabetic patients, shedding light on the challenges posed by these pathogens and highlighting the urgent need for concerted efforts in surveillance, prevention, and treatment to mitigate their impact on public health.
This is the prospective study conducted over a period of 12 months (January 2023 to December 2023). Institutional Ethical Committee (IEC) approval was obtained. This study consisted all non-duplicate n = 500 different clinical samples from diabetic patients which were received for bacterial culture in the microbiology department during the study period. This study included various clinical samples (n = 500) from diabetic patients that were received for bacterial culture in the microbiology department during the course of the study.
Microbiological analysis
After being incubated at 37 °C overnight, the isolated colony was grown in pure culture. The colony was then identified using a routine process that included evaluating the suspected colonies biochemically. Antibiotic susceptibility testing was done on these isolates using a Gram-negative antibiotic panel.6
Determination of antimicrobial susceptibility
The pattern of antimicrobial susceptibility was identified. They were evaluated for zones of inhibition in accordance with the CLSI guidelines. Amikacin, ampicillin, cefepime, cefazolin, ceftazidime, cefuroxime, cefotaxime, gentamicin, pipercillin-tazobactam, ciprofloxacin, co-trimoxazole, meropenem and imipenem were among the antibiotics that were tested and findings were compared with the ATCC strain.7
Detection of Drug-resistant K. pneumoniae
Based on the findings of the disc diffusion method, Multi Drug Resistant (MDR), Extensively Drug Resistant (XDR), and Pan Drug Resistant (PDR) strains were identified.
AmpC beta-lactamases
- Cefoxitin (30 µg) disk was used for screening.
- Following incubation, if the zone’s diameter is greater than 14 mm, cefoxitin may be a potential source of AmpC beta-lactamases.8,9
Double disk synergy test
- Third-generation cephalosporin-resistant bacterial isolates were tested for ESBL production.
- Cefotaxime (30 µg) at a distance of 15 mm from piperacillin/tazobactam edge to edge; incubated at 37 °C at 24 hours; zone of inhibition increased by more than 5 mm in diameter, indicating the presence of ESBL formation.10
Metallo-β-lactamase – MBL
- Combined disc test: imipenem (10 µg) and
0.1 M anhydrous EDTA (10 µl) were employed in one. - An increase in the zone diameter surrounding the EDTA disc of more than 5 mm is considered positive when compared to the imipenem disc.11
The process of identifying virulence factors
Hypermucoviscosity (HMV)
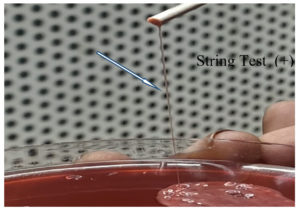
- The colony’s propensity to expand like a mucoviscous string
- A longer-than-10 mm string expansion is indicated of the HMV phenotype.
Figure 1 shows a hypervirulent strain of K. pneumoniae that passed the string test (Figure 2).12
Blood Hemolysis
- The isolates were placed in blood agar plates that had 5% sheep blood in order to perform the hemolysis test.
- After incubation for 24 hours at 37 °C, hemolysis was observed.13
Biofilm forming assay
- The biofilm productions were evaluated using the microtiter plate method. Klebsiella isolates attachment to an inert substrate was investigated.
- The strains were kept for 24 hours at 37 °C in Brain Heart Infusion Broth (BHIB). 48-well polystyrene microtiter plate with flat bottom wells was filled with 50 µl of the culture dilution, and it was incubated for 48 hours.
- Following incubation wells were gently washed three times using sterile saline and methanol fixation was carried out for 20 minutes.
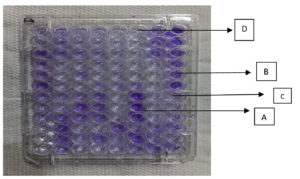
- Following a crystal violet stain, each well was washed. 1 ml of ethanol was used to decolorize the biofilm-associated crystal violet. The optical density was measured at 620 nm (Figure 3).14
Figure 3. Biofilm formation in microtitre plate assay
Note: A – Strong, B – Moderate, C – Weak, D – None
Extraction and amplification of 16S rRNA
DNA was extracted using DNA Extraction kit. Absorbance at 280 nm wavelengths was used to measure the concentration and purity of DNA.15
For the amplification of the 16S rRNA gene, universal primers were used. The reverse primer was R 5′-ACGGTTACCTTGTTACGACTT-3′, and the forward primer was F 5′-AGAGTTTGATCCTGGCTCAG-3′. 3 ml of template DNA, 2 ml of forward and reverse primer, 5.5 ml of DH2O, and 12.5 ml of master mix make up the total volume of a 25 ml PCR reaction mixture.16
The PCR technique was standardised to include an initial denaturation at 94 °C for
15 minutes, followed by 35 cycles of denaturation for 1 minute at 94 °C, annealing for 1 minute at 52 °C, extension for 1 minute and 30 seconds at 72 °C, and final extension for 5 minutes at 72 °C. With the amplified PCR sample, 1% Agarose gel electrophoresis was carried out using 1X TAE (Tris-Acetate-EDTA) buffer containing ethidium bromide.
The image was generated using a UV transilluminator while the amplified bands were visible using the gel documentation.17
Determination of multidrug-resistance genes
Specific primers and PCR reaction were used to identify the following genes: blaVIM, blaCTX-M, blaNDM, rmpA, WCaG, blaKPC, IMP, magA (K1), and wzy (K2). These genes are described in Table 1 and Table 2 provides the amplification conditions.
Table (1):
Primers for the identification of target genes
| PCR | Target gene | Sequences of Primer (5’-3’) | Product Size (bp) |
|---|---|---|---|
| Resistant genes | |||
| 1. | CTX-M | CGCTTTGCGATGTGCAG-ACCGCGATATCGTTGGT | 550 |
| 2. | NDM | GGGCAGTCGCTTCCAACGGT-GTAGTGCTCAGTGTCGGCAT | 476 |
| IMP | TTGACACTCCATTTACDG-GATYGAGAATTAAGCCACYCT | 139 | |
| VIM | GATGGTGTTTGGTCGCATA-CGAATGCGCAGCACCAG | 390 | |
| KPC | CATTCAAGGGCTTTCTTGCTGC-ACGACGGCATAGTCATTTGC | 538 | |
| Biofilm gene | |||
| 3. | wcaG | GGTTGGKTCAGCAATCGTA-ACTATTCCGCCAACTTTTGC | 169 |
| Hyper virulent gene | |||
| 4. | rmpA | ACT GGG CTA CCT CTG CTT CA-ACT GGG CTA CCT CTG CTT CA | 550 |
| 5. | magA (K1) | GGTGCTCTTTACATCATTGC-GCA ATG GCC ATT TGC GTT AG | 282 |
| 6. | Wzy (K2) | GACCCGATA TTC ATA CTT GAC AGA G-CCT GAA GTA AAA TCG TAA ATA GAT GGC | 641 |
Table (2):
PCR-Amplification conditions
PCR |
Initial Denaturation |
Denaturation |
Annealing |
Elongation |
Final Elongation |
Cycle |
|---|---|---|---|---|---|---|
1. |
95°C 7 min |
95 °C 1 min |
58 °C 1 min 30 sec |
72 °C 2 min |
72 °C 5 min |
40 cycles |
2. |
95°C 5min |
95 °C 1 min |
55 °C 2 min |
72 °C 1 min |
72 °C 7 min |
35 cycles |
3. |
95°C 10 min |
95 °C 1 min |
56 °C 1 min 45 sec |
72 °C 1 min |
72 °C 10 min |
45 cycles |
4. |
97°C 7 min |
97 °C 1 min |
59 °C 1 min 45 sec |
72 °C 2 min |
72 °C 7 min |
40 cycles |
5. |
95°C 10 min |
95 °C 1 min |
56 °C 1 min 30 sec |
72 °C 1 min |
72 °C 10 min |
45 cycles |
6. |
95°C 7 min |
95 °C 1 min |
52 °C 1 min 45 sec |
72 °C 2 min |
72 °C 7 min |
35 cycles |
In the current investigation, 500 non-duplicate clinical samples in diabetic patients were obtained from a tertiary care hospital. Among the 500 samples, 386 bacterial isolates were identified (386/500), 20.7% (n = 80/386) were in blood, 27.4% (n = 106/386) in urine, 14.5% (n = 56/386) in pus, 8% (n = 31/386) in stool, 15.2% (n = 59/386) in wound swab and 13.9% (n = 54/386) in sputum sample. Distribution of bacterial pathogens is shown in Table 3.
Table (3):
Distribution of pathogens obtained from clinical specimens
Source of the organism |
E. coli (%) |
Klebsiella sp. (%) |
A. baumanii (%) |
Pseudomonas sp. (%) |
Citrobacter sp. (%) |
Shigella sp. (%) |
Enterobacter sp. (%) |
Salmonella sp. (%) |
Proteus sp. (%) |
|---|---|---|---|---|---|---|---|---|---|
Blood |
10 |
22 |
12 |
4 |
– |
– |
– |
2 |
– |
Urine |
23 |
28 |
14 |
18 |
6 |
– |
7 |
– |
2 |
Pus |
4 |
27 |
9 |
15 |
6 |
– |
4 |
– |
– |
Stool |
7 |
13 |
– |
– |
– |
7 |
– |
2 |
– |
Wound swab |
6 |
48 |
14 |
17 |
2 |
– |
– |
– |
6 |
Sputum |
2 |
14 |
14 |
21 |
– |
– |
– |
– |
– |
Total |
52 |
152 |
63 |
75 |
14 |
7 |
11 |
4 |
8 |
The majority of these MDR K. pneumoniae isolates were found in wound swab/exudate samples (n = 21), urine (n = 11), pus (n = 14), and blood (n = 6). From a total of 152 non-duplicate isolates of K. pneumoniae, 68 isolates were identified as MDR and these were further included for characterization.
The majority of MDR isolates were found in males (n = 41), followed by females (n = 27). Klebsiella spp. were isolated more frequently from urine samples (n = 28; 34.47%), pus samples (n = 27; 17.37%), sputum samples (n = 14; 12.63%), blood samples (n = 22; 9.74%) and wound swabs (n = 48; 8.68%). The distribution analysis of Klebsiella spp. around clinical wards indicated that approximately n = 83; 36.6% isolates were from the OPD. Approximately (n = 48; 13%), (n = 21; 13%) of Klebsiella spp. were isolated from the IPD and ICU, respectively (Table 4). The majority of MDR Klebsiella isolates (36.7%) were collected from wounds, with pus (22%) and blood (19.1%), urine (13.2%), and sputum (6%). According to this, the majority of MDR K. pneumoniae isolates were obtained from IPD (n = 21), OPD (n = 46), and ICU (n = 13). Hospital-acquired infections (n = 8), community-acquired infections (n = 15), and diabetic infections (n = 59) were linked to the majority of MDR K. pneumoniae cases.
Table (4):
Occurrence of Klebsiella spp. in clinical sample
| Klebsiella spp. n = 152 | MDR Klebsiella spp. n = 68 | |
|---|---|---|
| Gender | ||
| Male | 93 | 41 |
| Female | 59 | 27 |
| Community-acquired Infections (CAIs) | 36 | 15 |
| Hospital-acquired Infections (HAIs) | 19 | 8 |
| Diabetic Infections | 97 | 59 |
| Clinical sources | ||
| Urine | 28 | 9 |
| Blood | 22 | 13 |
| Pus swab | 27 | 15 |
| Sputum | 14 | 6 |
| Wound swab | 48 | 25 |
| Hospital sites | ||
| ICU | 21 | 13 |
| IPD | 48 | 21 |
| OPD | 83 | 46 |
Note: IPD- in-patient department; OPD- out-patient department; ICU- intensive care unit
Resistance profile of Klebsiella pneumonia isolates
Table 5 represents the percentage of isolates exhibiting resistance toward thirteen antibiotics. During the study period ciprofloxacin, cefazolin, and Nitrofurantoin appeared to be the most effective drugs, as 93%, 92.1% and 90% of ESBL-producing K. pneumonia were screened. The highest resistance observed among MBL-producing K. pneumoniae were cefazolin (95.5%), followed by Nitrofurantoin (90.7%) and Tigecycline (83.8%). We found similar antibiotic resistance trends in MDR K. pneumoniae isolated from different clinical sources. Although the percentage of K. pneumoniae strains that were resistant to Meropenem and Imipenem was lower. A comparison between the resistance pattern of isolates from Diabetic infections versus community-acquired infections indicated similar resistance patterns toward the antibiotics mentioned in Table 5.
Table (5):
Antimicrobial resistance patterns of multidrug-resistance Klebsiella sp.
Antibiotics |
ESBL-producing Klebsiella pneumoniae (%) |
MBL-producing Klebsiella pneumoniae (%) |
|---|---|---|
Cefazolin |
92.6 |
95.5 |
Cefepime |
55.8 |
77.9 |
Imipenem |
7.3 |
11.7 |
Meropenem |
5.5 |
8.2 |
Ciprofloxacin |
92.1 |
80.7 |
Moxifloxacin |
72.0 |
67.6 |
Gentamycin |
64.7 |
61.7 |
Amikacin |
35.2 |
30.8 |
Nitrofurantoin |
90 |
90 |
Norfloxacin |
72 |
67 |
Tetracycline |
81 |
75 |
Chloramphenicol |
85.2 |
72 |
Citrimoxazoles |
75 |
80.8 |
Tigecycline |
90 |
83.8 |
A total of 68 MDR K.pneumoniae isolates were found to exhibit reduced susceptibility to meropenem and imipenem. These 68 isolates were further studied for ESBL and MBL production. Among ESBL positive isolates of K. pneumoniae, 53 isolates showed presence of blaSHV (n = 53, 77.9%), blaTEM (n = 51, 75%) and blaCTX-M (n = 42, 61.7%), blaTEM with blaSHV positive for 31 isolates, blaTEM with blaCTX-M positive for 27 isolates and 19 isolates were positive for blaTEM with blaSHV and blaCTX-M. ( 4) Among 32 MBL positive K. pneumoniae, blaKPC was positive for (n = 32, 47%), blaVIM + blaIMP (n = 31, 45.5%), blaVIM (n = 28, 41.1%), blaIMP (n = 24, 35.2%) and blaKPC + blaVIM (n = 23, 33.8%) were identified. Similarly, MBL resistant genes were also represented in Table 6.
Table (6):
Distribution of ESBL and MBL Resistant genes in Klebsiella pneumomiae
| MDR K. pneumoniae (n = 68) | |
|---|---|
| Name of Gene | n (%) |
| ESBL | |
| blaTEM alone | 51 |
| blaSHV alone | 53 |
| blaCTX-M alone | 42 |
| blaTEM + blaSHV | 31 |
| blaTEM + blaCTX-M | 27 |
| blaTEM + blaSHV + blaCTX-M | 19 |
| MBL | |
| blaKPC alone | 32 |
| blaVIM alone | 28 |
| blaIMP alone | 24 |
| blaOXA-48 alone | 26 |
| blaNDM alone | 18 |
| blaKPC + blaIMP | 23 |
| blaVIM + blaIMP | 31 |
| blaIMP + blaNDM | 17 |
| blaKPC + blaIMP | 21 |
| blaOXA-48 + blaNDM | 11 |
| blaKPC + blaVIM + blaIMP | 17 |
| blaKPC + blaOXA-48 + blaNDM | 19 |
| blaVIM + blaIMP + blaOXA-48 | 13 |
The increasing prevalence of antibiotic resistance is limiting the potential treatment choices for diseases caused by bacteria that have developed resistance to drugs. Greece has been shown to have the highest rate of carbapenem resistance globally, at 68%, followed by India and the eastern Mediterranean regions at 54%.18 Table 1 lists the most frequently reported ESBLs in India, which include SHV, TEM, and CTX-M. Manoharan et al. and Goyal et al. have documented co-existence of SHV, TEM, and CTX-M in Enterobacteriaceae, which is similar to this study’s observation.19,20 Study by Tsay et al. revealed that 49% of diabetes mellitus patients had bacteremia acquired in the community as a result of a K. pneumoniae infection.21
Gram-negative bacteria are more deadly than gram-positive bacteria and have been linked to infection in healthcare settings.22 North East India might be concerned about the increasing number of hospital-acquired and community-acquired multidrug-resistant bacterial infections. The current investigation also shows that the stains that were isolated from different clinical cases had a high level of resistance to all widely used antibiotics. As with previous studies from this region, the least amount of resistance was observed against imipenem, gentamicin, and amikacin.23 As a result of treatment with carbapenem, there is a correlation between increased production of ESBL 77.9% and carbapenem resistance.24 According to the current investigation, 11% of the bacteria had both ESBL and MBL genes, and 47% of the isolates were MBL producers. The increased incidence was similarly found by Devi et al.25 Multidrug-resistant pathogen colonization is becoming more prevalent in ICU patients due to their main illnesses, which frequently require recurrent hospital stays.26
Globally, the majority of cases are found in intensive care units.27 The majority of the time, ESBL development and antimicrobial resistance in Klebsiella were connected. The WHO classified ESBL-producing Klebsiella as highly pathogenic superbugs in 2017.28 Our isolates of K. pneumoniae showed multidrug-resistance, with an increase in carbapenem resistance. Imipenem and meropenem are the two medicines that are effective in treating CRKP. It may be advantageous to use a combination of antibiotics in along with removing invasive devices.29
In our research work, Klebsiella exhibited resistance to second- and third-generation cephalosporins. This shows the importance of screening gram-negative bacteria for cephalosporin resistance and the development of ESBL and MBL.30 Based on the genes, carbapenemases are considered to be the main mechanism that causes the development of CRKP isolates. In this study, blaKPC gene was present in majority, according to MBL resistance gene analysis. We identified a higher incidence of blaOXA-48 and no blaVIM in any of the isolates we examined, compared to another study by Ghaith et al.31 with a different genotypic profile. Regarding ESBL genes, 42 isolates received our investigation with blaCTX-M. In an investigation by Amer et al., similar isolates were found, which is in accordance with this observation.32
The isolates in this study were found to be detected with other carbapenemases, including blaKPC, blaVIM, and blaOXA-48. In bacteremic patients with MDR K. pneumoniae, showing indicates that a combination treatment may be associated with better prognosis than monotherapy alone.33 The emergence of multidrug resistance has usually been attributed to the rapid dissemination and fast spread of resistant Klebsiella spp. The hypervirulent strains of K. pneumoniae have emerged; these strains were initially predominant among isolates that were susceptible to antibiotics. They have recently been observed in MDR isolates, as the study also showed. This presents further treatment challenges because virulence associated with antibiotic resistance can be highly concerning, possibly resulting in extremely high rates of mortality.34 Hospital-acquired infections were more common than community-acquired infections, according to our overall analysis, but the higher isolation percentages from the OPD indicated an increased probability that the infections were diabetic infections.
In summary, there has been a rise in the frequency and antibiotic resistance of K. pneumoniae isolates. Antibiotic stewardship is necessary to prevent diabetic patients from overusing antibiotics, as K. pneumoniae represents a severe threat to the healthcare system. Strict infection control measures must be implemented in hospital settings. It can be challenging to diagnose and treat because of how different the indications and symptoms can be. The development of appropriate healthcare standards to combat AMR resistance and subsequently decrease the death rate requires a thorough understanding of the bacteria that cause infections, their antibiotic susceptibility profile, the specific cause of resistance, and the geographic pattern of resistance. The present study demonstrated that the isolates showed higher levels of drug resistance as well as greater prevalence of ESBL and MBL producers. The development of appropriate healthcare standards to combat AMR resistance requires a thorough understanding of the bacteria that cause infections, their antibiotic susceptibility profile, the precise cause of resistance, and the distribution pattern of resistance. The current investigation demonstrated an increase in drug resistance in the isolates as well as a higher frequency of ESBL and MBL producers. Effective therapy is currently being delayed because there are currently few available therapeutic options due to the increasing emergence of multidrug-resistant organisms. Developing protection and management methods should be used in hospitals. Information obtained from this study may be significant in developing new community treatment strategies and in providing baseline information that may be used to implement antibiotic guidelines that will decrease the development of drug resistance. To detect new MDR-Kp infections as soon as possible, infection management measures including antibiotic stewardship programs with continuous monitoring are essential.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Jagan V for invaluable suggestions and support during the course of our work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethical Committee, SRM Medical College Hospital and Research Centre, Chengalpattu, India, vide reference number 2937/IEC/2021.
- Ali MR, Yang Y, Dai Y, et al. Prevalence of multidrug-resistant hypervirulent Klebsiella pneumoniae without defined hypervirulent biomarkers in Anhui, China: A new dimension of hypervirulence. Front Microbiol. 2023;14:1247091.
Crossref - Chen L, Kreiswirth BN. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect Dis. 2018;18(1):2-3.
Crossref - Pattolath A, Adhikari P, Pai V. Carbapenemase-Producing Klebsiella pneumoniae Infections in Diabetic and Nondiabetic Hospitalized Patients. Cureus. 2024;16(1):e52468.
Crossref - Solgi H, Shahcheraghi F, Bolourchi N, Ahmadi A. Molecular characterization of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae ST11 harbouring blaNDM-1 and blaOXA-48 carbapenemases in Iran. Microb Pathog. 2020;149:104507.
Crossref - Shen D, Ma G, Li C, et al. Emergence of a multidrug-resistant hypervirulent Klebsiella pneumoniae sequence type 23 strain with a rare blaCTX-M-24 -harboring virulence plasmid. Antimicrob Agents Chemother. 2019;63(3):e02273-18.
Crossref - Kar B, Sharma M, Peter A, et al. Prevalence and molecular characterization of b-lactamase producers and fluoroquinolone resistant clinical isolates from North East India. J Infect Public Health. 2021;14(5):628-637.
Crossref - Das A, Sahoo RK, Gaur M, et al. Molecular prevalence of resistance determinants, virulence factors and capsular serotypes among colistin resistance carbapenemase producing Klebsiella pneumoniae: a multi-centric retrospective study. 3 Biotech. 2022;12(1):30.
Crossref - Ravi V, Vijay DD, Sujhithra A, Jayanthi S, Subramanian TK, Harish N. Neonatal Sepsis: The impact of Hypervirulent Klebsiella pneumonia in a Tertiary Care Hospital. J Pure Appl Microbiol. 2022;16(3):2035-2044.
Crossref - Younas S, Ejaz H, Zafar A, Ejaz A, Saleem R, Javed H. AmpC beta-lactamases in Klebsiella pneumoniae: An emerging threat to the paediatric patients. J Pak Med Assoc. 2018;68(6):893-897.
- Aminul P, Anwar S, Molla MM, Miah MR. Evaluation of antibiotic resistance patterns in clinical isolates of Klebsiella pneumoniae in Bangladesh. Biosaf Health. 2021;3(06):301-306.
Crossref - Dhungana K, Awal BK, Dhungel B, Sharma S, Banjara MR, Rijal KR. Detection of Klebsiella pneumoniae carbapenemase (KPC) and metallo betalactamae (MBL) producing Gram negative bacteria isolated from different clinical samples in a Transplant Center, Kathmandu, Nepal. Acta Scientific Microbiology. 2019;2(12):60-69.
Crossref - Shankar CS, Nabarro LE, Anandan SA, et al. Extremely high mortality rates in patients with carbapenem resistant, hypermucoviscous Klebsiella pneumoniae blood stream infections. J Assoc Physicians India. 2018;66(12):13-16.
- Das A, Behera BK, Acharya S, et al. Genetic diversity and multiple antibiotic resistance index study of bacterial pathogen, Klebsiella pneumoniae strains isolated from diseased Indian major carps. Folia Microbiol. 2019;64(6):875-887.
Crossref - Nirwati H, Sinanjung K, Fahrunissa F, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019;13(Suppl 11):20.
Crossref - Ghaffarian F, Hedayati M, Ebrahim-Saraie HS, Roushan ZA, Mojtahedi A. Molecular epidemiology of ESBL-producing Klebsiella pneumoniae isolates in intensive care units of a tertiary care hospital, North of Iran. Cell Mol Biol. 2018;64(7):75-79.
Crossref - He Y, Guo X, Xiang S, et al. Comparative analyses of phenotypic methods and 16S rRNA, khe, rpoB genes sequencing for identification of clinical isolates of Klebsiella pneumoniae. Antonie Van Leeuwenhoek. 2016;109(7):1029-1040.
Crossref - Singh SK, Mishra M, Sahoo M, et al. Antibiotic resistance determinants and clonal relationships among multidrug-resistant isolates of Klebsiella pneumoniae. Microb Pathog. 2017;110:31-36.
Crossref - World Health Organization. Antimicrobial resistance: global report on surveillance. World Health Organization. 2014.
- Manoharan A, Premalatha K, Chatterjee S, Mathai D, SARI Study Group. Correlation of TEM, SHV and CTX-M extended-spectrum beta lactamases among Enterobacteriaceae with their in vitro antimicrobial susceptibility. Indian J Med Microbiol. 2011;29(2):161-164.
Crossref - Goyal A, Prasad KN, Prasad A, Gupta S, Ghoshal U, Ayyagari A. Extended spectrum b-lactamases in Escherichia coli & Klebsiella pneumoniae & associated risk factors. Indian J Med Res. 2009;129(6):695-700.
- Tsay R, Siu LK, Fung C, Chang F. Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch Intern Med. 2002;162(9):1021-1027.
Crossref - Agyepong N, Govinden U, Owusu-Ofori A, Essack SY. Multidrug-resistant gram-negative bacterial infections in a teaching hospital in Ghana. Antimicrob Resist Infect Control. 2018;7:37.
Crossref - Gajamer VR, Bhattacharjee A, Paul D, et al. The first report of phenotypic and molecular characterization of extended-spectrum beta-lactamase-producing uropathogens in Sikkim and Darjeeling hills of India. Microb Drug Resist. 2018;24(9):1284-1288.
Crossref - Giriyapur RS, Nandihal NW, Krishna BVS, Patil AB, Chandrasekhar MR. Comparison of disc diffusion methods for the detection of extended-spectrum beta lactamase-producing Enterobacteriaceae. J Lab Physicians. 2011;3(1):33-36.
Crossref - Devi U, Bora R, Das JK, Mahanta J. Extended-spectrum b-lactamase & carbapenemase-producing gram-negative bacilli in neonates from a tertiary care Centre in Dibrugarh, Assam, India. Indian J Med Res. 2018;147(1):110-114.
Crossref - Kuo HY, Yang CM, Lin MF, Cheng WL, Tien N, Liou ML. Distribution of blaOXA carrying imipenem-resistant Acinetobacter spp. in 3 hospitals in Taiwan. Diagn Microbiol Infect Dis. 2010;66(2):195-199.
Crossref - Pillay D, Naidoo L, Swe-Han KS, Mahabeer Y. Neonatal sepsis in a tertiary unit in South Africa. BMC Infect Dis. 2021;21:1-10.
Crossref - Wang N, Zhan M, Liu J, et al. Prevalence of carbapenem-resistant Klebsiella pneumoniae infection in a Northern Province in China: clinical characteristics, drug resistance, and geographic distribution. Infect Drug Resist. 2022;15:569-579.
Crossref - Ibrahim ME, Abbas M, Al-Shahrai AM, Elamin BK. Phenotypic characterization and antibiotic resistance patterns of extended-spectrum b-Lactamase-and AmpC b-lactamase-producing Gram-negative bacteria in a referral hospital, Saudi Arabia. Can J Infect Dis Med Microbiol. 2019;2019(1):6054694.
Crossref - Bedzichowska A, Przekora J, Stapinska-Syniec A, et al. Frequency of infections caused by ESBL-producing bacteria in pediatric ward-single center five-year observation. Arch Med Sci. 2019;15(3):688-693.
Crossref - Ghaith D, Morsy SA, Sebak M, Rabea RA. Phenotypic and genotypic characterization of carbapenem-resistant Gram-negative organisms, Beni-Suef, Egypt. Beni-Suef Univ J Basic Appl Sci. 2023;12(1):61.
Crossref - Amer R, El-Baghdady K, Kamel I, El-Shishtawy H. Prevalence of extended spectrum Beta-Lactamase Genes among Escherichia coli and Klebsiella pneumoniae clinical isolates. Egypt J Microbiol. 2019;54(1):91-101.
Crossref - Qureshi ZA, Paterson DL, Potoski BA, et al. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob Agents Chemother. 2012;56(4):2108-2113.
Crossref - Shankar C, Nabarro LEB, Ragupathi NKD, et al. Draft genome sequences of three hypervirulent carbapenem-resistant Klebsiella pneumoniae isolates from bacteremia. Genome Announcements. 2016;4(6):10-128.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.