The ability to acquire iron from the environment is often an important virulence factor for pathogenic bacteria and Vibrios are no exception to this. Vibrios are reported mainly from marine habitats and most of the species are pathogenic. Among those, the pathogenic vibrios eg. V cholerae, V. parahaemolyticus, V. vulnificus causes foodborne illnesses. Vibrios are capable of producing all different classes of siderophores like hydroxamate (aerobactin), catecholate (vibriobactin, fluvibactin), carboxylate (vibrioferrin), and amphiphilic (amphibactin). Every different species of vibrios are capable of utilizing some endogenous or xenosiderophores. Being Gram-negative bacteria, Vibrios import iron siderophore via TonB dependent transport system and unlike other Gamma proteobacteria these usually possess two or even three partially redundant TonB systems for iron siderophore transport. Other than selected few iron siderophores, most pathogenic Vibrios are known to be able to utilize heme as the sole iron source, while some species are capable of importing free iron from the environment. As per the present knowledge, the spectrum of iron compound transport and utilization in Vibrios is better understood than the siderophore biosynthetic capability of individual species.
Iron Transport, Siderophore, Vibrios, Iron Acquisition, Ferrisiderophore
Iron plays a crucial role in various physiological processes of bacterial growth and pathogenesis. It is an essential yet limiting nutrient for all living organisms except for Borrelia burgdorferi.1 In the environment, iron exists in two interconvertible oxidation states: ferrous iron (Fe2+) present in the reducing environment and ferric iron (Fe3+) predominant in the oxidizing environment.2 The ability of human pathogenic bacteria to utilize iron is expected to be essential for the establishment of both an infection in their hosts as well as surviving in the host habitat. Bacteria take up iron majorly through secreted low molecular weight iron capturing peptides known as siderophores or by taking up preformed iron-loaded heme or by directly taking up soluble ferrous iron.2,3 The iron siderophore complex and other iron complexes uptake systems in Gram-negative bacteria are homologous, comprising specific receptors present on the outer membrane, periplasmic binding protein, cytoplasmic permease, and energy is derived by ATPase activity. Most bacteria require elemental iron to synthesize multiple essential enzymes, which are involved in important cellular processes, such as cytochromes for cellular metabolism, ribonucleotide reductase that is ultimately involved in the nucleic acid synthesis, and enzymes and cofactors for Kreb’s cycle.2 Iron has very low solubility in the water (⁓10-18 M) at natural physiological pH, the environment in which bacteria grow. Bacteria required a minimum iron concentration of ⁓10-6 M in the cytoplasm for normal physiological process.4
Vibrios require iron like most other organisms. A large spectrum of Vibrio species are pathogenic that cause several diseases like cholera, vibriosis, ear, and eye otitis. Vibrio cholerae mainly causes gastrointestinal diarrheal disease popularly known as Cholera. At the time of infection in humans, the cells adhere to the surface of epithelial cells of the small intestine and colonize, resulting in a heavy load of V. cholerae in the small intestine. Further, it secretes an enterotoxin, known as cholera toxin (CTX), that interferes with cAMP production in epithelial cells, causing a lot of water and ion efflux leading to diarrhea. Vibrio cholerae produces multiple iron chelating siderophores under low iron concentration or iron restriction conditions. These siderophore molecules frequently have catechols, carboxylates, hydroxamates, and amphiphilic groups, which help in iron acquisitions.5 These siderophore iron acquisition systems help V. cholerae to grow in low iron conditions, moreover, it has been observed that if V. cholerae is grown with siderophore producing bacteria, it results in higher proliferation of V. cholerae. In the presence of siderophore producing commensal E. coli, V. cholerae established better infection in adult mice. This study strongly recommended that V. cholerae may overcome environmental and host iron restrictions by using xenosiderophores during infection as well aquatic growing environments.6 Other than V. cholerae V. parahaemolyticus is also reported to cause acute gastroenteritis in humans,7 which is the most important enteropathogen found in Japan and Taiwan.8 Vibrio parahaemolyticus produces siderophore vibrioferrin, enterobactin, aerobactin, and ferrichrome. Another species, V. ordalii also causes vibriosis mainly in salmonid fishes, although the mechanism of virulence is not fully understood. V. ordalii produces siderophore piscibactin under iron limiting growth environment which is used for iron acquisition and as virulence factor.9 Fish pathogens have evolved various iron uptake systems, some of those iron uptake systems act as virulence factors. Heme and siderophore uptake are the most studied systems in fish pathogenesis. Among these, the Siderophore anguibactin or pscibactin from Vibrio and Photobacterium, have been reported as a virulence factor in fish pathogenesis.10 Vibrio alginolyticus, another halophilic marine bacterium tends to cause superficial skin infection resulting in wound and ear infection, otitis media, and otitis externa.11 Siderophores produced by Vibrio alginolyticus have been reported and biosynthesis of siderophores is affected by the gene luxO and rpoN but mechanism of iron uptake by siderophores are not fully understood.12 Outer membrane protein U (OmpU) is recently identified as a protein from V. alginolyticus, involved in iron balancing in vitro as disruption of ompU resulted in a 78% decrease in extracellular iron levels.13 Vibrio vulnificus, a highly virulent marine bacterium, cause serious wound infections and fatal septicemia.14 V. vulnificus produces siderophore vulnibactin, aerobactin, Desferoxamine B, and vibriobactin.15 V. vulnificus has three paralogs of the TonB iron transport systems named TonB1, TonB2, and TonB3 systems. A deletion mutant in tonB1 and tonB2 are defective in iron transport in V. vulnificus. These double mutants of V. vulnificus cultured in iron rich medium did not induce any significant changes in pathogenesis when studied in Caenorhabditis elegans model system.16 Vibrios have multiple iron transport mechanism that allows the maximum iron acquisition from their growing environment. Most of the iron uptake systems found in vibrios are diligently related, this relation indicates they are evolved from common genetic ancestor. Some iron transport systems are adopted through the horizontal gene transfer and may have niche specific iron transport roles in different vibrios.17 Genes for the iron homeostasis system are present on both chromosomes of Vibrio cholerae and encode ferric or ferrous iron transporter and siderophore specific receptors for various types of siderophore chelated iron and regulators. Few iron transport systems are well- characterized in Vibrio cholerae, those constitute more than 1% of their genome.17 As there are scanty reports on iron acquisition systems in Vibrio, therefore, this review gives a holistic view of molecular mechanisms involved in these.
Iron Acquisition
Iron acquisitions or transport systems recognize iron in the growing medium which is complexed with siderophore compounds known as iron chelators or iron carrier molecules.18 These molecules are either biologically synthesized by the bacteria for their own use or scavenged from a growing medium containing the exogenous source. Vibrio spp. synthesizes various types of siderophores that have a high affinity to ferric iron (Table 1). When these iron chelators named siderophores are secreted into the bacterial habitat or growing medium, they play an important role in iron acquisitions. The siderophores bind to the free iron and form ferric siderophore complexes, these complexes are recognized by specific receptors present on the bacterial outer membrane for further transport inside cell.3
Table (1):
Siderophore synthesis and utilization by Vibrio species.
Species |
Endogenous Siderophores |
Reference(s) |
Xenosiderophores |
Reference(s) |
|---|---|---|---|---|
Vibrio cholerae |
Vibriobactin |
31,76 |
Enterobactin Ferrichrome |
27,29 76 |
Vibrio parahaemolyticus |
Vibrioferrin |
77 |
Enterobactin Aerobactin Ferrichrome |
78 42 |
Vibrio vulnificus |
Vulnibactin Uncharacterized Hydroxamate |
79
15 |
Aerobactin Deferoxamine B Vibriobactin |
51
80, 81 82 |
Vibrio harveyi |
Amphi-enterobactin Uncharacterized Hydroxamate Anguibactin |
64
83 |
||
Vibrio alginolyticus |
Uncharacterized Hydroxamate |
84 |
Heme |
84 |
Vibrio mimicus |
Aerobactin |
85 |
||
Vibrio hollisae |
Aerobactin |
86 |
||
Vibrio furnissii |
Desferrioxamine B |
87 |
||
Vibrio fluvialis |
Fluvibactin |
88 |
||
Vibrio fischeri |
Aerobactin |
52 |
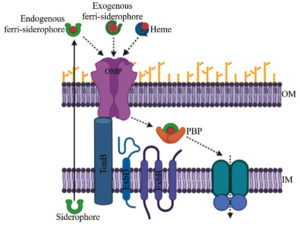
Acquisitions of iron from the environment to cytoplasm via an outer membrane and inner membrane require energy that is provided by TonB1, TonB2, and TonB3 systems. The TonB1 system has only three proteins known as TonB1, ExbB1, and ExbD1 for providing energy.19 TonB2 is very specific so that it uptakes only a few iron-bound substrates in V. vulnificus and V. cholerae.20-22 TonB2 system has total of six protein components TonB2, ExbB2, ExbD2, TtpB2, TtpC2, and TtpD2. All six components play an indispensable role in the acquisition of both endogenously produced siderophore iron complex as well xenosiderophore iron complex. TonB3 system is much more similar to the TonB2 system than the TonB1 system, it has possessed six components TonB3, ExbB3, ExbD3, TtpB3, TtpC3, and TtpD3.22,23 Ferrisiderophore complexes are first recognized by the outer membrane receptors which allow them to enter into the periplasmic space and bind to the periplasmic binding protein (PBP), after that, they are received by the inner membrane protein (IMP) for further transport to the cytoplasm.24 Outer membrane receptors are more sensitive or specific than periplasmic binding protein and inner membrane protein. In the cytoplasm, iron reduction and enzymatic cleavage are mandatory for the release of the iron to make available for the physiological process.17
Host iron containing molecules like transferrin, lactoferrin, and heme is generally utilized by the many pathogenic organisms. Some vibrios can use heme as a sole iron source for their growth.17 Besides these above mentioned iron compounds, ferrous and ferric iron are also utilized by the vibrios. Both the Iron siderophore and heme transport systems of Vibrio follow a similar multi-component mechanism (Fig. 1).
Ferric-siderophore Iron Transport Systems
Iron acquisition mechanism is well studied in both Gram-negative as well as in Gram-positive bacteria. Pathogenic bacteria have a diverse mechanism to overcome the limitation of iron in the host environment. The two most familiar mechanisms are the acquisition of iron with the help of siderophores and the utilization of iron from heme or other iron storage molecules in the human host. Both transport mechanisms are present in most pathogenic bacteria and the same is the case of Vibrio. Various components of iron siderophore and heme transport in Vibrio are presented in Table 2. A study suggested that these iron acquisition systems likely arise during the evolution of bacteria.25,26 Besides these two irons transport systems, vibrios also possess ferric and ferrous iron transport systems. In this review, we summarize the updated knowledge about some of the iron transport systems present in Vibrio spp.
Table (2):
Ferric-siderophore transport proteins by different Vibrio species.
| Siderophores | OMP | PBP | IMP | ATPase | References |
|---|---|---|---|---|---|
| Vibrio Cholerae | |||||
| Vibriobactin | ViuA | VctP | VctD, VctG | VctC | |
| Enterobactin | ViuA | VctP | VctD, VctG | VctC | 29 |
| Ferrichrome | FhuA | FhuD | FhuB | FhuC | 32 |
| Vibrio parahaemolyticus | |||||
| Vibrioferrin | PvuA | PvuB | PvuC,PvuD | PvuE | 39,78 |
| Enterobactin | ViuA | VctP | VctD,VctG | VctC | 30 |
| Aerobactin | IutA | VatD | VatB | VatC | 78 |
| Ferrichrome | FhuA | FhuD | FhuB | FhuC | 42 |
| Vibrio vulnificus | |||||
| Vulnibactin | VuuA | FatB,VatD | VuuB | PvuE | 36,37,80 |
| Aerobactin | IutA | VatD | VatB | VatC | 51 |
| Deferoxamine B | DesA | FhuD | FhuB | FhuC | 42,80,81 |
| Vibriobactin | ViuA | VctP | VctD, VctG | VctC | 80,90 |
| Vibrio harveyi | |||||
| Amphi-enterobactin | FapA | 64 | |||
| Anguibactin | FatA | FatB | FatC | FatD | 60 |
| Vibrio mimicus | |||||
| Aerobactin | IutA | FhuD | FhuB | FhuC | 85 |
| Vibrio hollisae | |||||
| Aerobactin | IutA | FhuD | FhuB | FhuC | 85,86 |
| Vibrio furnissii | |||||
| Desferrioxamine B | DesA | FhuD | FhuB | FhuC | 87 |
| Vibrio fluvialis | |||||
| Fluvibactin | ViuA/VctA | ViuP/VctP | ViuD/VctD | ViuCVctC | 88 |
| Vibrio fischeri | |||||
| Aerobactin | IutA | FhuD | FhuB | FhuC | 52,85,86 |
OMP- Outer Membrane Protein, PBP- Periplasmic Binding Protein, IMP- Inner Membrane Protein
ViuPDGC
ViuPDGC is the most important iron transport system which transports catechol type siderophores in Vibrio spp,27 also known as VctPDGC. It is responsible for the transport of the catechol type siderophores vibriobactin and enterobactin. These siderophores are transported through the membrane by PBP-dependent ABC transporter system VctPDGC or ViuPDGC. It also promotes the growth of E. coli enterobactin biosynthetic mutant in iron restriction conditions, and feoB, tonB, and aroB which are not required for this transportation.27
V. cholerae synthesizes and secretes vibriobactin, a catechol siderophore into the growing environment. An outer membrane receptor ViuA/VctA and periplasmic binding protein ViuP/VctP helps ABC transporters ViuABCD or VctABCD during the transportation of iron-vibriobactin siderophore complex.28,29 In this ABC transporter, a monomeric PBP transfers the ferrisiderophore to its inner membrane receptors. These inner membrane permeases consist of two fundamental membrane permeases and two copies of ATPase proteins which provide energy for the transport of iron vibriobactin complex.30 Vibrio cholerae also transport xenosiderophores that are not synthesized by themselves including catechol siderophore enterobactin and hydroxamate ferrichrome.31,32
The location of the vctPDGC genes is on chromosome 2 of Vibrio cholerae.29,33 Studies indicated that VctP/ ViuP is the PBP, VctD, and VctG are IMP proteins, and VctC is known as the ATPase. VctA is a TonB dependent OMP receptor for uptake of enterobactin siderophores.29 When the VctPDGC is introduced in fepB mutant of E. coli, it confirmed it to be a siderophore transporter. Gene fepB is the PBP, a component of the ABC transporter for the acquisition of enterobactin siderophores. The VctPDGC filling the gap which arises due to the fep mutation, also suggests that Vct transports the enterobactin.34 VctPDGC increases the transportation of catechol siderophore like ligands and promotes the growth of E. coli SM193w. VctPDGC genes promote the growth of E. coli in which EntF and TonB are mutated or absent. This indicates that VctPDGC can transport enterobactin independently through the outer membrane. In Vibrio cholerae, VctPDGC constitutes a functional iron siderophore complex transporter in the mutant of vib, feo and fbp.30 V. cholerae has various iron acquisition systems that provide the capability of bacteria to uptake of iron sources from different environments. The expression of iron acquisition systems is highly complicated and extremely regulated by the iron homeostasis regulator gene Fur. Recently, a protein named IurV synthesized by the ORF VC0231 reportedly play a key role in the regulation of the iron acquisition mechanism. A deletion in VC0231 resulted in upregulation of 52 genes and downregulation of 21 genes, most of the interfered genes are linked to the iron acquisition and iron homeostasis. This indicated that the,gene IurV is a new novel regulator of the AraC/XylS gene family, those represses iron uptake genes.35
VuuAB, FatB, and VatCD
Vulnibactin is a catechol type siderophore transported through the ferric-siderophore transport system comprised of VuuAB, FatB and VatCD. VuuA is a ferric-vulnibactin receptor protein that recognizes the iron vulnibactin complex from the environment. The vuuA gene in V. vulnificus is regulated by the Fur regulator.36,37 Mutation in vuuA results in loss of the utilization of ferric-vulnibactin as a sole source of iron and also their pathogenesis. VuuB, FatB, and VatD act as ferric-vulnibactin binding periplasmic proteins. VatC is the ATPase that provides energy to the system for the transportation of ferric-vulnibactin. Genes vuuA, vuuB and fatB are found in a single operon with three open reading frames of the length of 2058 bp, 816 bp, and 909 bp respectively.36 These open reading frames finally produce three proteins consisting of 685, 271, and 302 amino acid residues, having a molecular weight of 76, 30, and 34 kDa. Mutants of vuuA were unable to grow under iron limiting environments whereas wild type was grown normally which suggested that vuuA receptor plays an important role in the growth of V. vulnificus M2799.37 Double mutants of vuuB and fatB, also showed growth impairment but growth was restored after 6 hours of incubation. This indicated that V. vulnificus M2799 has another protein that complements the defective genes. VuuB, FAD containing siderophore interacting protein family play a cruicial role in ferric-vulnibactin transportation. IutB, belong to ferric-siderophore reductase family, which is a substitute of VuuB in the absence of VuuB. However, it is still not clearly understood why V. vulnificus M2799 has both VuuB and IutB transport proteins with the same functions.38 In silico analysis suggested that FatB is homologous to a PBP ferric-vulnibactin, VatD in V. vulnificus CMCP6 genome. The double mutant of fatB and fatD grow poorly than the single mutant under the iron limiting environments.36,37 These studies suggested that VuuA, VuuB, FatB, and VatD are essential for ferric-vulnibactin mediated iron acquisition in Vibrio vulnificus.
Fig. 2. Dendrogram obtained for Outer Membrane Proteins (OMP), showing evolutionary relationships among them
PvuBCDE
Vibrio parahaemolyticus produces a siderophore vibrioferrin belonging to the family polyhydroxycarboxylate. It is transported across the plasma membrane with the help of the ferric-vibrioferrin transport system PvuBCDE. These four genes pvuBCDE present in an operon containing nine genes, four out of nine genes named pvsABDE are responsible for vibrioferrin synthesis and pvsC is responsible for the putative exporter. The genes pvuBCDE, encode for proteins showing strong homology to E. coli FecBCDE, respectively. These iron transport genes are transcribed as polycistronic mRNA with the outer membrane receptor gene pvuA and biosynthetic gene psuA.39 The gene pvuA encodes 78 kDa ferric vibrioferrin receptor protein, the sequence of which displays 31% identity and 48% similarity to RumA, a receptor for rhizoferrin in Morganella morganii.40 PvuA has 712 amino acid residues with a molecular mass of the mature protein 75.080 kDa which is almost equal to 78 kDa estimated by SDS-PAGE with isoelectric points 4.71.40 Mutation in gene pvuA results in loss of the ability to transport ferric-vibrioferrin, whereas complementation of pvuA restores the ferric-vibrioferrin acquisition.
Nucleotide sequencing of pvuBCDE indicates four complete ORFs within the same transcript, those are designated as pvuB, pvuC, pvuD and pvuE. The starting codon of pvuB is far away approximately 101 bp from the end of the preceding functional gene pvuA, there is no promoter present in the pvuB gene. The PvuB protein sequence displays 49% identity and 66% similarity to the PBP FecB of E. coli. PvuC and PvuD have been identified as cytoplasmic inner membrane permease, this is homology to FecC with 38% identity and 55% similarity. The PvuC and PvuD are also homologous to FecD of E. coli with 43% identity and 63% similarity, respectively. PvuC and PvuD have also shown homology with each other, suggesting that PvuC and PvuD proteins make heterodimers for membrane-localized transport complex in V. parahaemolyticus. The amino acid sequences of PvuE display strong homology with FecE of E. coli (56% identity and 75% similarity), ATP binding protein of ABC transport system.39 Deletion of ∆vp0057 from wild type V. parahaemolyticus potentially changes the function of VP0057, proteins related to iron transport and T6SS1 are upregulated in ∆vp0057 strains. The vp0057 deletion promotes the acquisition of Fe2+ and Fe3+ which is regulated by the iron-related and T6SS1 related proteins respectively.41
FhuABCD and DesA
FhuABCD found in most of the pathogenic bacteria including E. coli and Vibrio spp, is responsible for the transportation of ferrichrome. Ferrichrome is a hydroxamate type of siderophore transported through ATP-dependent transporter FhuABCD. Transportation of this iron ferrichrome complex across both membranes in Gram-negative bacteria is mediated by multiple proteins. Among those, FhuA is present in the outer membrane to recognize the ferrichrome siderophores. In this iron transport system, the periplasmic binding protein is FhuD, cytoplasmic membrane permease is FhuB, and ATPase, FhuC associated with the inner side of the cytoplasmic membrane. Desferrioxamine B is recognized by the outer membrane protein DesA, then follows the FhuABCD system same as ferrichrome for transportation.42-44
This Iron transport system FhuABCD has been studied in detail for E. coli K-12 for ferrichrome transport. It is encoded by a single operon system consisting of genes fhuA, fhuB, fhuC and fhuD.45 The outer membrane protein receptor is encoded by the fhuA gene that recognizes the ferrichrome siderophores for uptake. The FhuA protein forms a cylindrical channel in the outer membrane that is closed most of the time and is opened when needed for ferrichrome transport, the energy provided by the TonB, ExbB, and ExbD.44,46,47 The opening and closing of FhuA are determined by a loop formed from residue 316 to residue 356. Deletion of the loop converts the protein into a permanently open outer membrane channel.46 FhuA recognizes the ferrichrome and after the internalization, ferrichrome binds to the periplasmic binding protein FhuD and is transferred from FhuD to the cytoplasmic membrane permease protein FhuB.48,49 Transmembrane protein, FhuB is twice larger than the other ABC transporter inner membrane permease proteins composed of two separate polypeptides. The separation of both polypeptides of FhuB gives rise to two halves FhuB(N) and FhuB(C) and both can reconstitute at cytoplasmic membrane for the transport of ferrichrome.48 Two copies of the ATPase FhuC interact with FhuB(N) and FhuB(C). FhuD binds to ferrichrome and brings that to FhuB channel. Mutation in FhuD results in a reduced binding capacity with FhuB.50
VatABCD and IutA
VatABCD is responsible for the transportation of siderophores aerobactin in Vibrio spp. Corresponding genes of the system are found in an operon, the first unit encoding an operon containing three genes (vatCDB) and the third unit having a single gene (iutA), that encodes a 76 kD ferric aerobactin receptor, homologous to IutA. The second unit also has another single gene (iutR) that encodes transcriptional repressors homologous to GntR family. Expression of the first and third unit of the operon is iron regulated but the expression of the second unit is not influenced by iron concentration present in the growing medium.51 The protein product of vatCDB having amino acid sequences similar to ABC transporter component for ferric aerobactin in V. mimicus MatB, C and D and in E. coli FhuB, C and D. Study suggested that VatC acts as an ATP binding protein, VatD plays a role of periplasmic binding protein and VatB is designated as an inner membrane protein. The expression of iutA gene from Vibrio spp. gives rise to a protein of 718 amino acids protein, homologous to V. mimicus IutA and E. coli IutA. A signal sequence of 28 amino acids is present in the 76 kDa precursor product of iutA outer membrane protein. VatCDB operon present in close vicinity to gene iutA recommended that VatCDB transporter is specific only to ferric aerobactin. Disruption of vatD of V. vulnificus results in loss of the ability to utilize ferric aerobactin as a xenosiderophores, indicating that VatCDB operon responsible for ferric aerobactin transport. Production of IutA increased independently to the absence of aerobactin when the mutation was introduced in iutR gene. In wild-type strains, low-level production of IutA in the lack of aerobactin under an iron limiting environment was observed. This suggested that iutA promoter is fully active only when an iron restricted environment in addition to the expression of aerobactin.51 V. fischeri ES114 negatively interferes with the growth of other Vibrio spp. The aerobactin exporter gene aerE and transcriptional activator of aerobactin production are dependent on the expression gene LuxT. Under an iron restriction environment, aerobactin production helps V. fischeri ES114 to exclude other Vibrio species that do not have aerobactin production genes. V. fischeri ES114 having a mutation in aerobactin production gene loss competition with other Vibrio spp. Insertion of iutA with fhuCDBb into V. harveyi, results in aerobactin cheater lifestyle.52
FatDCBAE
Transport system FatDCBAE is solely responsible for the transport of ferric-anguibactin complexes. Ferric-anguibactin is transported into the periplasmic space through a barrel-shaped outer membrane receptor FatA specific for ferric-anguibactin, transported into periplasmic space.53,54 Outer membrane proteins acquire energy by the TonB system and ATP is generated by the proton motive force on cytoplasmic membrane.55,56 Three TonB systems, TonB1, TonB2, and TonB3 are reported in Vibrio vulnificus TonB2 system is responsible for ferric-anguibactin transport and virulence of V. harveyi. TonB2 and TonB3 systems have six components of protein TonB2, ExbB2, ExbD2, TtpB2, TtpC2, TtpD2 and TonB3, ExbB3, ExbD3, TtpB3, TtpC3, TtpD3 respectively.23 Besides the TonB system proteins, another protein TtpC is also reported for ferric siderophore transport through the TonB2 system. The TtpC proteins are found in all vibrios and play an indispensable role in TonB2 system, therefore, facilitating ferric siderophore transport in Vibrio spp.20-22,57 After the recognition of ferric-anguibactin by the FatA, ferric-anguibactin complex is transferred to the PBP FatB, and finally, ferric-anguibactin is received by the heterodimer of FatC and FatD inner membrane permease proteins.58 FatB, FatC, and FatD play a very essential role in the ferric-anguibatin transportation in Vibrio species. Besides FatBCD, two ATPase FvtE and FatE are involved in the transportation of ferric-anguibactin complex. The fvtE is a part of operon coding for ABC siderophore transporter including FatBCD.59,60 Some studies showed that fvtB, fvtC and fvtD are only specific for the transportation of ferric-vanchrobactin but not for ferric-anguibactin. The pJM1 encoded ABC transporter FatBCD-FatE and ABC transporter FvtBDCE encoded by the chromosome are specific for transportation of two siderophore complex ferric-anguibactin and ferric-vanchrobactin. The interesting observation is FvtE can also transport the ferric-anguibactin complex. 36,60-63
FapA
FapA is a ferric-siderophore specific outer membrane receptor protein that recognizes the amphi-enterobactin siderophores. FapA protein is present in the fraction of outer membrane proteins of Vibrio campbellii HY01, and is very crucial for the transportation of complex ferric-amphi-enterobactin. Amphi-enterobactin is a siderophore like enterobactin, where fatty acid is attached to enterobactin or enterobactin similar molecules. The amphi-enterobactin biosynthetic gene cluster contains six genes aebABCDEF that encode the proteins that are homologs to enterobactin biosynthetic genes cluster.64,65 The amphi-enterobactin siderophore gene cluster of V. harveyi contains a gene named fapA, homologous to ferric-siderophore specific outer membrane protein gene, specific for transportation of ferric-amphi-enterobactin.65 Studies did not give sufficient information on whether amphi-enterobactin siderophore biosynthetic genes are present in various specific strains or widespread in Vibrio compbellii and Vibrio harveyi strains. In silico analysis of the sequences around amphi-enterobactin biosynthetic gene cluster revealed that fapA is located just downstream vicinity of the aebD gene. The strong homology of FapA proteins between both BAA-1116 and HY01 strains of V. compbellii has been found. FapA has also a similarity to ferric-siderophore specific outer membrane proteins of E. coli FepA, The transport system of amphi-enterobactin has not been fully known and understood till now.65
HutABCD for Heme iron transport
Besides ferric-siderophore iron transport systems, vibrios have more than one receptor for the transport of heme iron. Acquisition of heme iron in Vibrio cholerae is dependent on three heme iron receptors, designated as HutA, HutR, and HasR.66 These receptors allow transporting heme iron from the growing medium, containing heme as the sole source of iron during bacterial growth. Heme is transported into the bacterial cell in the form of an iron-porphyrin complex and supports heme biosynthetic mutants for growth.66 It is not completely understood how V. cholerae utilize heme; solely as whole heme complex or it is broken for utilization after uptake into the cytoplasm. The cytoplasmic protein HutZ, an enzyme similar to the heme-oxygenase enzyme to degrade heme, is required for the utilization of heme as a source of iron in Vibrio cholerae.67 Heme receptors HupA, HvtA, and HupO are also found in other species of vibrios such as V fluvialis68; and MhuA in V. mimicus.69 Operon system organization for both HutA and HutR is the same and closely related in V. cholerae.66,68 The ability to utilize hemoglobin as an iron source is less common in V. cholerae as compared to other species including V. parahaemolyticus, V. fluvialis, V. alginolyticus, V. vulnificus which can use hemoglobin more efficiently. hupA and hvtA are hemin receptors found in V. vulnificus, hupA is highly upregulated in elevation of temperature in the human pathogenic strains while hvtA is upregulated in lower temperature. Both hupA and hutA hemin receptors are involved in optimal hemin uptake in V. vulnificus, their expression is dually regulated by the concentration of iron present in the growing environment and temperature.70
HutA is an outer membrane receptor protein for the recognition of heme iron in V. cholerae; besides this, it has two additional outer membrane receptor proteins HutR and HasR. HutR has shown significant homology to HutA and other outer membrane receptor proteins present in Vibrio spp.66 A triple mutant for HutA, HutR, and HasR receptors results in a complete loss of utilization of heme as a sole source of iron. Heme iron receptors HutA and HutR of V. cholerae worked with one of the TonB systems either TonB1 or TonB2 but efficiently worked with TonB1 systems, whereas HasR is TonB2 dependent. Utilization of heme is more efficient when uptake is through HutA in presence of TonB1. HutB is a periplasmic binding protein that receives the heme iron from HutA, HutR, and HasR outer membrane receptors and delivers to inner membrane permease HutC and HutD in the case of V. cholerae.17,66,71 A gene HutZ found in V. cholerae, is needed for optimal utilization of transported heme. The hutZ is found in an operon with the other two genes hutW and hutX, transcribed from the TonB1 operon system. Poor growth of hutZ mutant in the medium where heme is used as the sole source of iron indicates a failure to efficiently utilize heme iron from the medium. Restoration of expression of hutWXZ genes in the mutants is reported to result in the utilization of heme iron as a sole source of iron.67
The energy required during heme transport through the outer membrane is generated by the bacterial proton motive force utilizing the ATPase. Components of the TonB system TonB, ExbB, and ExbD, interacting with TonB dependent outer membrane transporter helps in transfer of heme across the inner membrane by an ATP binding cassette transporter.72
FeoABC and FbpABC
Siderophores are having an extremely high affinity for iron and stimulate growth under iron limiting environments nevertheless their biosynthesis is energy consuming. This gap is filled by an additional system i.e. ferric iron and ferrous iron transport system, many carrier proteins taking part in this process are identified in the vibrios. The ferrous iron transport system FeoABC is distributed in all Vibrio species.73,74 This system is one of the earliest known bacterial iron transport systems. Homologs of the feo genes are generally present in bacterial species which could be due to the presence of Fe2+ as the predominant iron source during early evolution, The Feo proteins, FeoA, FeoB, and FeoC from V. cholerae function in equal molar concentration in vivo. FeoA and FeoC did not affect the rate of hydrolyzing NTPase but are essential for the Feo mediated iron transport.75
A plasmid constructed with deletion of feoABC in E. coli H1771 makes the strain unable to transport iron from the medium and resulting in growth inhibition. Another plasmid complemented with each of the mutations, inserted into the cell, restored the growth. These studies recommended that each feo gene is required for transportation of Fe2+ in V. cholerae.73 In the pathogenic V. cholerae, the feo operon has three genes feoA, feoB and feoC. The feoB is inner membrane permease, encoding 83 kDa protein having amino-terminal GTPase domain, form the ferrous iron transport channel. FeoA and FeoC are approximately having 8.5 kDa cytoplasmic membrane proteins, attached to the N-terminal cytoplasmic domain of FeoB and serve as secondary functions for FeoB.73
Besides the ferrous iron transport, V. cholerae has a ferric (Fe3+) iron transport system, FbpABC, an ABC transporter.73 This system consists of three genes fbpA, fbpB, and fbpC in a single operon system. Periplasmic binding protein FbpA is encoded by fbpA, inner membrane permease FbpB is encoded by fbpB and FbpC ATPase component is encoded by fbpC.73 Fur box is present upstream of fbpA and regulation of fbpA mRNA by Fur is observed. But fbpA and fbpC were also transcribed separately and not regulated by Fur. It was projected that ferric iron bind to outer membrane protein FbpA, then transferred to the periplasmic binding protein, and finally be handed off to FbpBC in the cytoplasmic membrane with the help of ATP delivered to the cytoplasm. Apart from this transport system, TonB independent system has also been reported by Fbp system. The Fbp iron transport system in vibrios has no specific outer membrane receptor for ferric iron. FbpABC iron transport system is highly conserved within vibrios and represents a standard mechanism of ferric iron acquisition.3
Survival of Vibrio species without iron acquisition is not possible as iron plays a key role in metabolism and acts as a regulator for many genes. Iron homeostasis is a very important regulation for most bacteria. Various species inhabits a huge variety of environmental niche and utilize a variety of iron acquisition mechanisms depending upon the surrounding environments and availability of iron e.g. ferric or ferrous, insoluble hydroxide, heme, and iron-sulfur cluster present in host cells or surrounding environments. Vibrios have evolved the mechanism of iron acquisition in all bioavailable forms of irons. Some studies also suggested that horizontal gene transfer may be responsible for the distribution of iron transport systems from one species to another species. Siderophore transport and utilization of heme, both mechanisms are commonly present in most of the Vibrio species, though there is still a knowledge gap about iron transport and regulation. The mechanism of iron acquisition is highly regulated to maintain the required concentration of iron inside the bacterial cells. When we looked at the phylogenetic relationship between the 12 known iron siderophore TonB dependent transporter outer membrane proteins of Vibrio, there were 3 clear clusters that have been found (see Fig. 2). Sequences of the outer membrane transporters were processed in CLUSTALW2 to make the dendrogram based on sequence similarity judged by alignment. The first cluster consisted of catecholate and carboxylate type siderophore transporters ViuA, VuuA, FvtA, and PvuA. The second cluster consisted of hydroxamate and catechol type siderophore FatA, DesA, and FhuA. The third cluster consisted of mostly heme transporters HutA, HutR, HasR, and aerobactin transporter IutA and enterobactin transporter FetA. The transporters may have been repurposed for their ligand affinity over the course of their evolution in Vibrio.
The vibrios can synthesize and utilize a great variety of iron-binding siderophores in iron limited environment. The utilization of siderophore is more diverse than synthesis indicates that some of the Vibrio species utilized those siderophores not synthesized by themselves, this type of siderophore is known as xenosiderophores. Siderophore vibriobactin is synthesized by V. cholerae but it can utilize enterobactin and ferrichrome as well. Every siderophore requires a specific transport system that includes outer membrane protein, periplasmic binding protein, inner membrane protein, TonB complex, and a cognate ATPase. Besides the siderophore iron uptake system, some vibrios can uptake heme iron. The heme iron uptake system is a member of TonB dependent iron transporter family and homologous to the ferri-siderophore iron uptake system. Vibrios have an additional iron uptake system FeoABC. This system is one of the earliest known bacterial iron acquisition systems. Vibrios are capable to survive nutrient poor marine environments and can rapidly adapt to new environmental conditions for their growth.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both the authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by the DST Science and Engineering Research Board, India with grant number ECR/2016/000630.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
Not applicable.
- Williams LR, Austin FE. Hemolytic activity of Borrelia burgdorferi. Infect Immun. 1992;60(8):3224-3230.
Crossref - Kenneth NR, Emily AD. Biochemical and Physical Properties of Siderophore. In: Crosa JH, Mey AR, Payne SM, eds. Iron Transport in Bacteria. ASM Press; 2004:3-17.
Crossref - Wyckoff EE, Mey AR, Payne SM. Iron acquisition in Vibrio cholerae. BioMetals. 2007;20(3-4):405-416.
Crossref - Lemos ML, Osorio CR. Iron Uptake in Vibrio and Aeromonas. Iron Uptake Homeost Microorg. 2010:117-141. https://www.caister.com/iron
- Biosca EG, Fouz B, Alcaide E, Amaro C. Siderophore-mediated iron acquisition mechanisms in Vibrio vulnificus biotype 2. Appl Environ Microbiol. 1996;62(3):928-935.
Crossref - Byun H, Jung IJ, Chen J, Valencia JL, Zhu J. Siderophore piracy enhances vibrio cholerae environmental survival and pathogenesis. Microbiol. 2020;166(11):1038-1046.
Crossref - Kaper JB, Morris JG, Levine MM. Cholera. Clin Microbiol Rev. 1995;8(1):48-86.
Crossref - Chiou AS, Chen L, Chen S-K. Foodborne Illness in Taiwan, 1981-1989. Food Aust. 1991;43(2):70-71.
Crossref - Ruiz P, Balado M, Fuentes-monteverde JC, et al. The Fish Pathogen Vibrio ordalii Under Iron Deprivation Produces the Siderophore Piscibactin. Microorgansms. 2019;7(9):313.
Crossref - Lemos ML, Balado M. Iron uptake mechanisms as key virulence factors in bacterial fish pathogens. J Appl Microbiol. 2020;129(1):104-115.
Crossref - Pezzlo M, Valter PJ, Burns MJ. Wound infection associated with Vibrio alginolyticus. Am J Clin Pathol. 1979;71(4):476-478.
Crossref - Wang Q, Liu Q, Ma Y, Rui H, Zhang Y. LuxO controls extracellular protease, haemolytic activities and siderophore production in fish pathogen Vibrio alginolyticus. J Appl Microbiol. 2007;103(5):1525-1534.
Crossref - Lv T, Dai F, Zhuang Q, et al. Outer membrane protein OmpU is related to iron balance in Vibrio alginolyticus. Microbiol Res. 2020;230:126350.
Crossref - Chuang YC, Yuan CY, Liu CY, Lan CK, Huang AHM. Vibrio vulnificus Infection in Taiwan : Report of 28 Cases and Review of Clinical Manifestations and Treatment. Clinical Infectious diseases. 1992;15(2):271-276.
Crossref - Simpson LM, Oliver JD. Siderophore production by Vibrio vulnificus. Infect Immun. 1983;41(2):644-649.
Crossref - Bowles AK, Wayne DJ, Kenton RJ. Vibrio vulnificus iron transport mutant has normal pathogenicity in C. elegans. µP microPublication Biol. 2019.
Crossref - Payne SM, Mey AR, Wyckoss EE. Vibrio Iron Transport : Evolutionary Adaptation to Life in Multiple Environments. Microbiol Mol Biol Rev. 2016;80(1):69-90.
Crossref - Neilands JB. Microbial Iron compounds. Annu Rev Biochem. 1981;50:715-731.
Crossref - Occhino DA, Wyckoff EE, Henderson DP, Wrona TJ, Payne SM. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol Microbiol. 1998;29(6):1493-1507.
Crossref - Kuehl CJ, Crosa JH. The TonB energy transduction systems in Vibrio species. Rev Futur Microbiol. 2010;5(9):1403-1412.
Crossref - Kustusch RJ, Kuehl CJ, Crosa JH. The ttpC Gene Is Contained in Two of Three TonB Systems in the Human Pathogen Vibrio vulnificus, but Only One Is Active in Iron Transport and Virulence. J Bacteriol. 2012;194(12):3250-3259.
Crossref - Stork M, Otto BR, Crosa JH. A novel protein, TtpC, is a required component of the TonB2 complex for specific iron transport in the pathogens Vibrio anguillarum and Vibrio cholerae. J Bacteriol. 2007;189(5):1803-1815.
Crossref - Barnes AD, Pfeifer HJ, Zbylicki BR, et al. Two novel proteins, TtpB2 and TtpD2, are essential for iron transport in the TonB2 system of Vibrio vulnificus. Microbiology open. 2020;9(1):e00947.
Crossref - Braun V, Hantke K. Recent insights into iron import by bacteria. Curr Opin Chem Biol. 2011;15(2):328-334.
Crossref - Miethke M, Marahiel MA. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol Mol Biol Rev. 2007;71(3):413-451.
Crossref - Chatterjee A, O’Brian MR. Rapid evolution of a bacterial iron acquisition system. Mol Microbiol. 2018;108(1):90-100.
Crossref - Wyckoff EE, Allred BE, Raymond KN, Payne SM. Catechol Siderophore Transport by Vibrio cholerae. J Bacteriol. 2015;197(17):2840-2849.
Crossref - Wyckoff EE, Valle A, Smith SL, Payne SM. A Multifunctional ATP-Binding Cassette Transporter System from Vibrio cholerae Transports Vibriobactin and Enterobactin. J Bacteriol. 1999;181(24):7588-7596.
Crossref - Mey AR, Wyckoff EE, Oglesby AG, Rab E, Taylor RK, Payne SM. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect Immun. 2002;70(7):3419-3426.
Crossref - Wyckoff EE, Payne SM. The Vibrio cholerae VctPDGC system transports catechol siderophores and a siderophore-free iron ligand. Mol Microbiol. 2011;81(6):1446-1458.
Crossref - Griffiths GL, Sigelj SP, Paynej SM, Neilandssll JB. Vibriobactin , a Siderophore from Vibrio cholerae. J Biol Chem. 1984;259(1):383-385.
Crossref - Rogers MB, Sexton JA, DeCastro GJ, Calderwood SB. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J Bacteriol. 2000;182(8):2350-2353.
Crossref - Heidelberg JF, Elsen JA, Nelson WC, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477-483.
Crossref - Elkins MF, Earhart CF. Nucleotide sequence and regulation of the Escherichia coli gene for ferrienterobactin transport protein FepB. J Bacteriol. 1989;171(10):5443-5451.
Crossref - Sachman-Ruiz B, Ibarra JA, Estrada-De Los Santos P, et al. IurV, encoded by ORF VCA0231, is involved in the regulation of iron Uptake Genes in Vibrio cholerae. Genes (Basel). 2020;11(10):1184.
Crossref - Kawano H, Miyamoto K, Sakaguchi I, et al. Role of periplasmic binding proteins, FatB and VatD, in the vulnibactin utilization system of Vibrio vulnificus M2799. Microb Pathog. 2013;65:73-81.
Crossref - Webster AC, Litwin CM. Cloning and characterization of vuuA, a gene encoding the Vibrio vulnificus ferric vulnibactin receptor. Infect Immun. 2000;68(2):526-534.
Crossref - Okai N, Miyamoto K, Tomoo K, et al. VuuB and IutB reduce ferric-vulnibactin in Vibrio vulnificus M2799. BioMetals. 2020;33(4-5):187-200.
Crossref - Tanabe T, Funahashi T, Nakao H, Miyoshi SI, Shinoda S, Yamamoto S. Identification and Characterization of Genes Required for Biosynthesis and Transport of the Siderophore Vibrioferrin in Vibrio parahaemolyticus. J Bacteriol. 2003;185(23):6938-6949.
Crossref - Funahashi T, Moriya K, Uemura S, et al. Identification and characterization of pvuA, a gene encoding the ferric vibrioferrin receptor protein in Vibrio parahaemolyticus. J Bacteriol. 2002;184(4):936-946.
Crossref - Ye C, Ge Y, Zhang Y, et al. Deletion of vp0057, a Gene Encoding a Ser/Thr Protein Kinase, Impacts the Proteome and Promotes Iron Uptake and Competitive Advantage in Vibrio parahaemolyticus. J Proteome Res. 2021;20(1):250-260.
Crossref - Funahashi T, Tanabe T, Shiuchi K, Nakao H, Yamamoto S. Identification and characterization of genes required for utilization of desferri-ferrichrome and aerobactin in Vibrio parahaemolyticus. Biol Pharm Bull. 2009;32(3):359-365.
Crossref - Burkhardt R, Braun V. Nucleotide sequence of the fhuC and fhuD genes involved in iron (III) hydroxamate transport: Domains in FhuC homologous to ATP-binding proteins. Mol Gen Genet. 1987;209(1):49-55.
Crossref - Killmann H, Benz R, Braun V. Properties of the FhuA channel in the Escherichia coli outer membrane after deletion of FhuA portions within and outside the predicted gating loop. J Bacteriol. 1996;178(23):6913-6920.
Crossref - Fecker L, Braun V. Cloning and expression of the fhu genes involved in iron(III)-hydroxamate uptake by Escherichia coli. J Bacteriol. 1983;156(3):1301-1314.
Crossref - Killmann H, Benz R, Braun V. Conversion of the FhuA transport protein into a diffusion channel through the outer membrane of Escherichia coli. EMBO J. 1993;12(8):3007-3016.
Crossref - Braun V. Energy-coupled transport and signal transduction through the Gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16(4):295-307.
Crossref - Koster W, Braun V. Iron(III) hydroxamate transport of Escherichia coli: Restoration of iron supply by coexpression of the N- and C-terminal halves of the cytoplasmic membrane protein FhuB cloned on separate plasmids. Mol Gen Genet. 1990;223(3):379-384.
Crossref - Rohrbach MR, Braun V, Koster W. Ferrichrome transport in Escherichia coli K-12: Altered substrate specificity of mutated periplasmic FhuD and interaction of FhuD with the integral membrane protein FhuB. J Bacteriol. 1995;177(24):7186-7193.
Crossref - Mademidis A, Killmann H, Kraas W, Flechsler I, Jung G, Braun V. ATP-dependent ferric hydroxamate transport system in Escherichia coli: periplasmic FhuD interacts with a periplasmic and with a transmembrane/cytoplasmic region of the integral membrane protein FhuB, as revealed by competitive peptide mapping. Mol Microbiol. 1997;26(5):1109-1123.
Crossref - Tanabe T, Naka A, Aso H, et al. A novel aerobactin utilization cluster in Vibrio vulnificus with a gene involved in the transcription regulation of the iutA homologue. Microbiol Immunol. 2005;49(9):823-834.
Crossref - Eickhoff MJ, Bassler BL. Vibrio fischeri siderophore production drives competitive exclusion during dual-species growth. Mol Microbiol. 2020;114(2):244-261.
Crossref - Lopez CS, Crosa JH. Characterization of ferric-anguibactin transport in Vibrio anguillarum. BioMetals. 2007;20(3-4):393-403.
Crossref - Lopez CS, Alice AF, Chakraborty R, Crosa JH. Identification of amino acid residues required for ferric-anguibactin transport in the outer-membrane receptor FatA of Vibrio anguillarum. Microbiol. 2007;153(2):570-584.
Crossref - Chu BCH, Vogel HJ. A structural and functional analysis of type III periplasmic and substrate binding proteins: Their role in bacterial siderophore and heme transport. Biol Chem. 2011;392(1-2):39-52.
Crossref - Postle K, Larsen RA. TonB-dependent energy transduction between outer and cytoplasmic membranes. BioMetals. 2007;20(3-4):453-465.
Crossref - Kustusch RJ, Kuehl CJ, Crosa JH. Power plays : iron transport and energy transduction in pathogenic vibrios. BioMetals. 2011;24(3):559-566.
Crossref - Actis LA, Tolmasky ME, Crosa LM, Crosa JH. Characterization and regulation of the expression of FatB, an iron transport protein encoded by the pJM1 virulence plasmid. Mol Microbiol. 1995;17(1):197-204.
Crossref - Naka H, Liu M, Crosa JH. Two ABC transporter systems participate in siderophore transport in the marine pathogen Vibrio anguillarum 775 (pJM1). FEMS Microbiol Lett. 2013;341(2):79-86.
Crossref - Naka H, Actis LA, Crosa JH. The anguibactin biosynthesis and transport genes are encoded in the chromosome of Vibrio harveyi: A possible evolutionary origin for the pJM1 plasmid-encoded system of Vibrio anguillarum? MicrobiologyOpen. 2013;2(1):182-194.
Crossref - Balado M, Osorio CR, Lemos ML. FvtA Is the Receptor for the Siderophore Vanchrobactin in Vibrio anguillarum: Utility as a Route of Entry for Vanchrobactin Analogues. Appl Environ Microbiol. 2009;75(9):2775-2783.
Crossref - Naka H, Crosa JH. Identification and characterization of a novel outer membrane protein receptor FetA for ferric enterobactin transport in Vibrio anguillarum 775 (pJM1). BioMetals. 2012;25(1):125-133.
Crossref - Bay L, Larsen JL, Leisner JJ. Distribution of three genes involved in the PJM1 iron-sequestering system in various Vibrio anguillarum serogroups. Syst Appl Microbiol. 2007;30(2):85-92.
Crossref - Zane HK, Naka H, Rosconi F, Sandy M, Haygood MG, Butler A. Biosynthesis of Amphi-enterobactin Siderophores by Vibrio harveyi BAA-1116 : Identification of a Bifunctional Nonribosomal Peptide Synthetase Condensation Domain. J Am Chem Soc. 2014;136(15):5615-5618.
Crossref - Naka H, Reitz ZL, Jelowicki AL, Butler A, Haygood MG. Amphi enterobactin commonly produced among Vibrio campbellii and Vibrio harveyi strains can be taken up by a novel outer membrane protein FapA that also can transport canonical Fe(III)-enterobactin. J Biol Inorg Chem. 2018;23(7):1009-1022.
Crossref - Mey AR, Payne SM. Haem utilization in Vibrio cholerae involves multiple tonB-dependent haem receptors. Mol Microbiol. 2001;42(3):835-849.
Crossref - Wyckoff EE, Schmitt M, Wilks A, Payne SM. HutZ is required for efficient heme utilization in vibrio cholerae. J Bacteriol. 2004;186(13):4142-4151.
Crossref - Datta S, Crosa JH. Identification and characterization of a novel outer membrane protein receptor required for hemin utilization in Vibrio vulnificus. BioMetals. 2012;25(2):275-283.
Crossref - Tanabe T, Funahashi T, Moon YH, Tamai E, Yamamoto S. Identification and characterization of a Vibrio mimicus gene encoding the heme/hemoglobin receptor. Microbiol Immunol. 2010;54(10):606-617.
Crossref - Datta S, Kenton RJ. Characterization of temperature-dependent hemin uptake receptors HupA and HvtA in Vibrio vulnificus. Microbiology Open. 2019;8(10):e905.
Crossref - Lemos ML, Osorio CR. Heme, an iron supply for vibrios pathogenic for fish. BioMetals. 2007;20(3-4):615-626.
Crossref - Wandersman C, Delepelaire P. Haemophore functions revisited. Mol Microbiol. 2012;85(4):618-631.
Crossref - Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J Bacteriol. 2006;188(18):6515-6523.
Crossref - Weaver EA, Wyckoff EE, Mey AR, Morrison R, Payne SM. FeoA and FeoC Are Essential Components of the Vibrio cholerae Ferrous Iron Uptake System , and FeoC Interacts with FeoB. J Bacteriol. 2013;195(21):4826-4835.
Crossref - Gomez-Garzon C, Payne SM. Vibrio cholerae FeoB hydrolyzes ATP and GTP in vitro in the absence of stimulatory factors. Metallomics. 2020;12(12):2065-2074.
Crossref - Keating TA, Marshall CG, Walsh CT. Vibriobactin Biosynthesis in Vibrio cholerae : VibH Is an Amide Synthase Homologous to Nonribosomal Peptide Synthetase Condensation Domains†. Biochem 2000;39(50):15513-15521.
Crossref - Yamamoto S, Okujo N, Yoshida T, Matsuura S, Shinoda S. Structure and Iron Transport Activity of Vibrioferrin, a New Siderophore of Vibrio parahaemolyticus. J Biochem. 1994;115(5):868-874.
Crossref - Funahashi T, Tanabe T, Aso H, et al. An iron-regulated gene required for utilization of aerobactin as an exogenous siderophore in Vibrio parahaemolyticus. Microbiology. 2003;149(5):1217-1225.
Crossref - Okujo N, Saito M, Yamamoto S, Yoshida T, Miyoshi S, Shinoda S. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals. 1994;7(2):109-116.
Crossref - Aso H, Miyoshi SI, Nakao H, Okamoto K, Yamamoto S. Induction of an outer membrane protein of 78 kDa in Vibrio vulnificus cultured in the presence of desferrioxamine B under iron-limiting conditions. FEMS Microbiol Lett. 2002;212(1):65-70.
Crossref - Choon-Mee K, Yong-Jin P, Sung-Heui S. A Widespread Deferoxamine-Mediated Iron-Uptake System in Vibrio vulnificus. J Infect Dis. 2007;196(10):1537-1545.
Crossref - Holmstrom K, Gram L. Elucidation of the Vibrio anguillarum genetic response to the potential fish probiont Pseudomonas fluorescens AH2, using RNA-arbitrarily primed PCR. J Bacteriol. 2003;185(3):831-842.
Crossref - Murugappan RM, Aravinth A, Karthikeyan M. Chemical and structural characterization of hydroxamate siderophore produced by marine Vibrio harveyi. J Ind Microbiol Biotechnol. 2011;38(2):265-273.
Crossref - Wang Q, Liu Q, Cao X, Yang M, Zhang Y. Characterization of two TonB systems in marine fish pathogen Vibrio alginolyticus: Their roles in iron utilization and virulence. Arch Microbiol. 2008;190(5):595-603.
Crossref - Moon YH, Tanabe T, Funahashi T, Shiuchi KI, Nakao H, Yamamoto S. Identification and characterization of two contiguous operons required for aerobactin transport and biosynthesis in Vibrio mimicus. Microbiol Immunol. 2004;48(5):389-398.
Crossref - Suzuki K, Tanabe T, Moon YH, et al. Identification and transcriptional organization of aerobactin transport and biosynthesis cluster genes of Vibrio hollisae. Res Microbiol. 2006;157(8):730-740.
Crossref - Tanabe T, Funahashi T, Miyamoto K, Tsujibo H, Yamamoto S. Identification of genes, desR and desA, required for utilization of desferrioxamine B as a xenosiderophore in Vibrio furnissii. Biol Pharm Bull. 2011;34(4):570-574.
Crossref - Yamamoto S, Okujo N, Fujita Y, Saito M, Yoshida T, Shinoda S. Structures of two polyamine-containing catecholate siderophores from vibrio fluvialis. J Biochem. 1993;113(5):538-544.
Crossref - Stoebner JA, Butterton JR, Calderwood SB, Payne SM. Identification of the Vibriobactin Receptor of Vibrio cholerae. J Bacteriol. 1992;174(10):3270-3274.
Crossref - Litwin CM, Rayback TW, Skinner J. Role of catechol siderophore synthesis in Vibrio vulnificus virulence. Infect Immun. 1996;64(7):2834-2838.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.