ISSN: 0973-7510
E-ISSN: 2581-690X
The aim of this present study to was to detect the 16S rRNA gene sequences of Escherichia coli local isolate from dialysis patients and to compare their genetic relatedness utilizing phylogenetic analysis. A total of (75) clinical samples (Urine) were collected from in dialysis patients but found thirty specimens of E. coli admitted at Baghdad Medical City, Baghdad Teaching Hospital during the period from October, 2017 to January, 2018 platinum calibrated sterile wire loop to transfer 1ml of the uncentrifuged urine specimen and streaked on MacConkey agar incubated at 37°C for 18-24 hours. The results of phylogenetic analysis showed that Proximity and the genetic dimension among themselves and the world with more than 98% to 99%compatibility values.
E. coli, UTI, 16S rRNA gene
Clinically, urinary tract infections (UTIs) are classified as uncomplicated or complicated; Complicated UTIs are known as UTIs associated with agents that compromise the urinary tract or steward defence, involving urinary obstruction, urinary retention raised by neurological disease, immunosuppression, renal failure, renal transplantation, pregnancy and the presence of foreign bodies such as calculi, indwelling catheters or another drainage devices (Flores-Mireles et al., 2015). Urinary tract contagion caused by bacteria associated with an increased antimicrobial resistant types are common in all age groups. Most urinary tract infections are initiated by organisms that gain entrance of the natural environment to the bladder through the urethra and are more prevalent in women than men, Urinary pathogens vary depending upon age, sex, catheterization, hospitalization and previous exposure for antimicrobials (Biadglegne and Abera, 2016). Escherichia coli is facultative anaerobe, Gram-negative bacilli, belong to Enterobacteriaceae. The lower intestine of human is the natural habitat of this bacterium, which resident in commensalism manner. However pathogenic Escherichia coli strains play a critical role in severe infections to human being (Farrokh et al., 2012). E. coli gives rise to maximal recurrent infections occur in human such as urinary tract infections, intestinal diseases, neonatal meningitis and septicemia (Motayo BO1 et al., 2012). In this study aim to the determine the role of 16S rRNA genes in detection of E. coli isolates in dialysis patients.
Samples Collection
A total of (75) clinical samples (Urine) were collected from in dialysis patients but found thirty specimens of E. coli admitted at Baghdad Medical City/ Baghdad Teaching Hospital during the period from October, 2017 to January, 2018.
Isolation and Identification of Escherichia coli
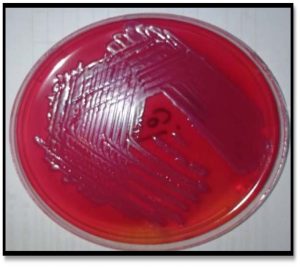
According to the diagnostic procedures recommended by MacFaddin and Benson (McFadden, 2000, Benson., 2001)Urine specimens were collected from the patients at admission to the hospital in a sterile container. A loopful of uncentrifuged urine samples was cultured onto blood agar, MacConkey agar and EMB media incubated for 24 hrs at 37°C, Fig. 1.
DNA extraction
Genomic DNA was extracted from clinical isolates of E. coli using (DNA mini kit that was supplied by G- spin DNA extraction kit, Korea) according to manufacturer’s instructions, primer were used in this study were obtained from microgen company (16S rRNA) Forward strand primers 5’- AGA GTTT GAT CCT GGC TCAG -3’ and Reverse strand primers 5’- CTT GTG CGGG CCCCC GTC AATTC -3’, Fig. 2.
PCR Amplification
The 16S rRNA gene was amplified using primer F (5’- AGA GTTT GAT CCT GGC TCAG -3’) and primer R (5’- CTT GTG CGGG CCCCC GTC AATTC -3’). The PCR amplification is performed in a total volume of 25ml containing 1.5ml DNA, 5ml Master Mix PCR (intron, korea), 1ml of each primer 10 pmol then nuclease-free water is added into a tube to a total volume from 25ml. Thermo cycling conditions were as follows: initial denaturation at 3 min at 95°C, followed by 35 cycles of denaturation 95°C for 45 sec, annealing at 62°C for 45sec, extension at 72°C for 45 sec and a final extension of 72°C for 7 min. The PCR products were separated on 2% agarose gel. The gel is left to run for 90 min with a 70 volt/65Am current. Following electrophoresis, visualization was conducted with a UV transilluminator after red stain staining.
Sequencing
The sequencing of 16S rRNA gene was performed at Macrogen Inc., by usingNCBI BLAST.
16S rRNA Genetic Polymorphism
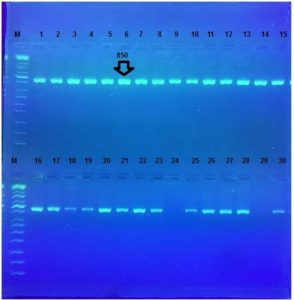
One and half µl of genomic DNA were used for each PCR reaction. A conventional PCR protocol was used to analyze simultaneously the presence of 16S rRNA gene. The presence of the 16S rRNA gene was identified by 850bp, Fig. 3.
Fig. 3. Agarose gel electrophoresis for16S rRNA gene (850bp). Bands were fractionated by electrophoresis on a 2% agarose gel (2 h., 5V/cm, 1X TBE) and visualized under U.V. light after staining with red stain. Lane: 1 (M: 100bp ladder)
The sequencing of amplified product of 16S rRNA gene for Study Groups
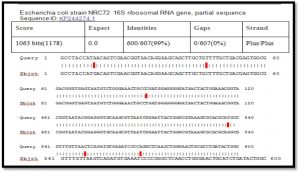
The sequencing of amplified product of 16S rRNA gene by direct sequencing. Our sequences were compared with the reference sequence of in national center biotechnology information (NCBI) Gene Bank.
A- G>C , T>A , A>T , G>A , T>G ,and G>T genotyping.
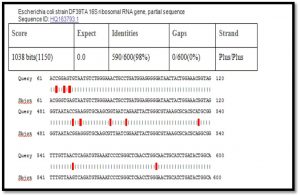
After alignment of product amplification of 16S rRNA gene for isolate 1 having two Transition G>A, eight Transversion G>C , T>A , A>T , T>G, and G>T from the Gene Bank, found that part of 16S rRNA gene having 98% compatibility with standard in Gene Bank 16S rRNA gene as shown in Table (1) (Singh SK.,2009).
B- G>A , C>T, T>C, G>T, and G>C genotyping.
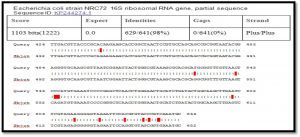
After alignment of product amplification of 16S rRNA gene for isolate 2 having four Transition G>A, C>T, T>C , three Transversion G>T, and G>C from the Gene Bank, found that part of 16S rRNA gene having 99% compatibility with standard in Gene Bank 16S rRNA gene, Table 2.
C- G>C , G>T, A>C, and G>A genotyping.
After alignment of product amplification of 16S rRNA gene for isolate 3 having three Transition G>A , nine Transversion G>C , G>T, A>C from the Gene Bank, found that part of 16S rRNA gene having 98% compatibility with standard in Gene Bank 16S rRNA gene, Table 3 (Ramesh et al., 2015).
D- M(A,C)>A ,and G>A genotyping.
After alignment of product amplification of 16S rRNA gene for isolate 4 having two Transition G>A, one Transversion M(A,C)>A from the Gene Bank, found that part of 16S rRNA gene having 99% compatibility with standard in Gene Bank 16S rRNA gene, Table 4 (Kohl KD et al.,2018).
Phylogenetic tree structuring
The phylogenetic tree diagrammatic by Molecular Evolutionary Genetics Analysis (MEGA) software version 6.0. Sequences that showed the highest identity (>98%) and maximum coverage (>99%), by comparison between isolated from dialysis patients Neighbor-joining tree was constructed for phylogenetic analysis. The genetic dimension between Iraq and the isolates of the world is detailed according to the Phylogenetic tree and the comparison table. Hierarchical cluster analysis determine the following clusters: large Cluster divided into several neck: first root the USA: Iraq and India the genetic dimension was by 2.3 it is close to UNITED KINGDOM ‘’ID: FN356960.1’’, third root including Iraq isolate E.coli the genetic dimension was by 2.7 it is close to Bangladesh ‘’ID: MG857757.1’’. The last cluster is divided into two branches the first branch Iraq the genetic dimension was by 3.5 it is close to Sweden “ID: CP029579.1”
When comparison between Escherichia coli isolated from dialysis patients recorded in the National Center Biotechnology Information (NCBI) and isolated from different source have under sequence (ID: HQ163793.1, ID: EU420950.1, ID: MF179674.1, ID: KY655125.1, ID: FN356960.1, ID: KP789332.1, ID: MG857757.1, ID: LM997164.1, ID: CP029973.1, ID: CP029579.1, ID: CP026473.1) (Shahi et al., 2016, Wang, C et al.,2008, Farag et al.,2017, Bozcal et al.,2018, Ogue et al.,2009, Momtaz et al.,2017, Johansen et al.,2014, Zurfluh et al.,2017, Kim et al.,2017) respectively with source of isolation and showed compatibility the highest identity (>98%) and maximum coverage (>99%) and score (1038-1216), and expect 0.0 with gene bank.
Submission of local Iraq isolate in NCBI
The 16S rRNA gene were registered after the correspondence of the National Center for Biotechnology Information and obtained accession number and became a reference to Iraq and the Middle East and the world. Ongoing work will add to this set as more type strains are publishedand it is available for download at NCBI: https://www.ncbi.nlm.nih.gov/nuccore/ MH628651.1, MH628665.1, MH628664.1, MH 628646.1.
The efficiency of genetic methods in the detection of accurate and rapid bacterial isolates proved near the Iraqi isolates E. coli of isolates Sweden
- Flores-Mireles A. L., Walker J. N., Caparon, M. & Hultgren S. J. Urinary Tract Infections: Epidemiology, Mechanisms Of Infection And Treatment Options. Nature Reviews Microbiology, 2015; 13, 269.
- Biadglegne F. & Abera B. Antimicrobial Resistance Of Bacterial Isolates From Urinary Tract Infections At Felge Hiwot Referral Hospital, Ethiopia. The Ethiopian Journal Of Health Development (EJHD), 2016; 23.
- Farrokh C, Jordan K, Auvray F, Glass K, Oppegaard H, Raynaud S,et al. Review Of Shiga-Toxin Producing Escherichia coli (STEC) And Their Significance In Dairy Production”. International Journal Of Food Microbiology., 2012; 162: 190-212.
- Motayo BO1, Ogiogwa IJ, Okerentugba PO, Innocent-Adiele HC, Nwanze, JC, Onoh CC, et al. Antimicrobial Resistance Profile Of Extra-Intestinal Escherichia coli Infections In A South Western Nigerian City.” Journal Of Microbiology Research, 2012; 2(5): 141-144.
- McFadden JF. Biochemical tests for identification of medical bacteria (3ed), Lippincott Williams and wilkins, USA. Microbiol. Rev., 2000; 2: 15-38.
- Benson HJ. Microbiological application, laboratory manual in general microbiology. 8th ed. W.b. Mcgraw Hill, 2001.
- Singh SK, Gupta K, Tiwari S, Shahi SK, Kumar S, Kumar A, and Gupta SK.Detecting aerobic bacterial diversity in patients with diabetic foot wounds using ERIC-PCR: a preliminary communication. Int J. Low Extrem Wounds.,2009; 8(4):203-8.
- Ramesh,N., Ramkumar S., Prasanth M., Gothandam K.M., Karthikeyan,S. and Bulent,B. Characterization of carbapenem-resistant Gram negative bacteria from Tamil Nadu. Characterization of carbapenem-resistant Gram-negative bacteria from Tamil Nadu Ramesh Nachimuthu, Ramkumar Subramani, Suresh Maray, K M Gothandam, Karthikeyan. Journal of chemotherapy (Florence, Italy), 2015.
- Kohl KD, Oakeson KF, Orr TJ, Miller AW, Forbey JS, Phillips CD, Dale C, Weiss RB, Dearing MD. Metagenomic sequencing provides insights into microbial detoxification in the guts of small mammalian herbivores (Neotoma spp.). FEMS Microbiol Ecol., 2018; 1;94(12).
- Shahi,S.K. and Kumar,A. A study on the relation of the severity of diabetic foot ulcers with the type of bacterial flora isolated from the wounds. International Surgery Journal, 2016; 3(1) P:189.
- Wang,C. and Dang,H. Incidence of ESBLs and plasmid-mediated AmpC beta-lactamase genesand integrons in environmental Enterobacteriaceae isolated from the eutrophic Jiaozhou Bay, China. World J Microbiol Biotechnol., 2008; 24:2889–2896.
- Farag M.M., Hassan M.M., Alzahrani,A.K., Abdallah,K.F., Ismail,A.K., Ganai,F.A. and Abdel-Moneim,A.S. Molecular subtyping and virulence markers of Escherichia coli isolates from Saudi Arabia, 2017.
- Bozcal E., Eldem V., Aydemir S. and Skurnik, M. Investigation of New Genomic Islands Related to Virulence in Extra intestinal pathogenic Escherichia coli (ExPEC) strains by in silico. Comparative Genomics Peer J., 2018; 24;6:e5470.
- Ogue E., Collins J., Gibson G.R. and Rastall R.A. Analysis of the antimicrobial effect of a synbiotic mixture containing Bifidobacterium bifidum 02 450B and galacto-oligosaccharides on the proteome of Clostridium perfringens, 2009.
- Momtaz F. and Majlish A.N.K. Molecular characterization of multi-drug resistant, adhesin producing alpha-hemolytic E. coli isolates causing recurrent urinary tract infections (RUTIs) in women in the Eastern part of Bangladesh. J Infect Dev Ctries, 2017; 11(6):459-469.
- Johansen,Jostein. and Haugum,Kjersti. NTNU, IKMM, Erling Skjalgs onsgate 1, 7006Trondheim, Norway, 2014.
- Zurfluh,K., Stevens M.J., Perterhans,S. and Stephan,R. 2017.Complete Genome Sequence of Escherichia coliABWA45, an rmtB-Encoding Wastewater Isolate. Genome Announc., 24; 5(34): e00844-17.
- Kim S, Jeong H, Kim EY, Kim JF, Lee SY, Yoon SH. Genomic and transcriptomic landscape of Escherichia coli BL21(DE3). Nucleic Acids Res., 2017; 45(9), 5285-5293.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.