ISSN: 0973-7510
E-ISSN: 2581-690X
House flies (Musca domestica L.) and green flies (Chrysomya megachepala L.) are two species of flies that are vectors of many types of bacteria in humans and animals throughout the world. The research was conducted to determine the sequence of the 16S rRNA gene and thus to conduct the phylogenic position, the bacteria isolated from Musca domestica and Chrysomya megachepala L used the 16S rRNA gene as a barcode. The bacteria identified were bacteria that have been tested to produce antibiotics. Pure cultures of each of the seven bacterial isolates were used for DNA analysis dan molecular identification. The results showed that the S1 isolate had a 90% similarity with the Bifidobacterium minimum [NR 044692.2]. Isolate S2, showing similarity of 86% with Alcaligenes faecalis [CP021883.1]. Isolate S3 has similarty 94% Brucella melitensis bv. [NR_076080.2]. Isolate S4 has a similarity of 99% with Sphingobacterium sp. [LK931720.1]. Isolate S5 has a similarity of 100% with Bacillus sp. [MH071158.1]. Isolate S6 has a similarity of 97% with Gamma proteobacterium [KJ127178.1]. While isolate S7 has a similarity of 100% Enterobacter asburiae [MH071322.1]. Species of bacteria identified from green flies and house flies that inhabit the city of Luwuk have never been reported to be associated with flies.
Bacteria, 16S rRNA, Musca domestica L., Chrysomya megachepala L., Luwuk City
The housefly, is one of the most common insects found worldwide. House flies (Musca domestica L.) and green flies (Chrysomya megachepala L.) are the two species of flies most commonly found around residential areas, rubbish bins, hospitals and other dirty areas in the tropics. In a previous study, morphological and genetic characteristics of the house fly and green fly from Luwuk City were distinguished, based on the cytochrome oxidase gene subunit 1 (CO1)1. The activity of house flies and green flies in dirty areas causes the two species of flies to become mechanical vectors of pathogenic microbial transmission.
All live stadium of house flies (e.g. larvae, pupae, adults) were commonly contaminated with a lot of kinds of microorganisms. However, adult house flies are highly mobile, they transport bacteria from septic environments to other substrates via contamination of their surfaces (legs, wings, bodies) and also by regurgitation of crop contents2,3. Bacteria on the internal and external parts of house flies vary greatly depending on geographical location and habitat4,5. Since house flies have the potential to become vectors of various types of pathogenic bacteria. The bacteria found in the larval stage of the fly are very different from the bacteria isolated from adult flies6. However, there are similarities in the types of bacteria in the internal parts of flies that live in different geographical areas6. Previous studies have identified bacterial isolates capable of producing antibiotic compounds against other bacteria in vitro tests. These bacteria were identified based on the 16S rRNA gene in this study. Molecular identification and metagenomic analysis of bacteria are very important to get antibiotic-resistant bacterial species and bacteria that cannot be isolated and cultured on artificial media7,8.
House flies have implications in the transmission of serious diseases such as anthrax, ophthalmia, typhoid fever, tuberculosis, cholera, and infantile diarrhea and transmit other pathogenic bacteria4,7. House flies can transmit more than 64 species of pathogenic bacteria in humans and animals, especially pathogenic microbes in the human digestive tract19,10. Musca domestica and C. megacephala are two species of flies that live in human settlements. Both species of flies have evolved with various species of pathogenic bacteria in humans, so it is very potential to transmit new species of pathogenic bacteria in humans1,12. Furthermore, C. megacephala is able to transmit 11-12 times more bacterial colonies than Musca domestica11,13.
House flies potential mechanical or biological vectors for the dissemination of pathogenic and multidrug-resistant bacteria8,15,16. In spite of these pathogenic microbes do not infect house flies. House flies have developed self-defense mechanisms against pathogenic microbes associated with the mouth organ, digestive tract and body surface9,17. Researchers from Macquarie University found antibiotics produced by microbes associated with flies, at each stage of fly development18. It should be suspected that house flies play a role in spreading antibiotic-resistant bacteria3,19,20. For this reason, research into the identification of bacteria from flies in various places in the world needs to be done continuously.
Isolation and identification of bacteria in house flies from various geographical locations is important to solve the types of pathogenic bacteria associated with flies. In this study, flies were obtained from the city of Luwuk. Luwuk is a coastal city in Central Sulawesi which, according to data from the Central Sulawesi Provincial Health Office in 2018, has a high digestive tract pathogenic infection. The location of the fly sample was obtained from several characteristics of the fly’s habitat, namely the traditional market area, residential areas, and hospitals. This research was conducted to identify molecular species of bacterial isolates from flies from various habitats in the city of Luwuk. The bacteria identified are bacteria that in previous studies have been known to produce antibiotics against other pathogenic bacteria.
Samples
Flies samples were taken from two locations in the city of Luwuk namely hospital waste dumps and traditional market waste dumps. Flies are caught with insect catching nets, directly preserved in sterile bottles sampels. Based on morphological identification, house flies and green flies were preserved separately in bottles.
Isolate bacteria from flies
Isolation of bacteria from wings of flies
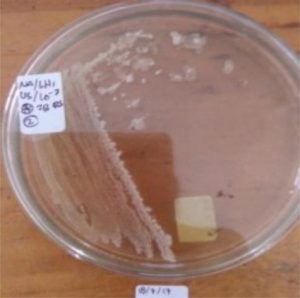
The flies were preserved with sterile bottles samples before being used as a source of bacterial inoculum, preserved again at sterile tube 60 minutes After 60 minutes, the fly is transferred to a sterile petri dish with a filter paper mat to remove residue on the surface of the fly’s body. Next, the flies were put into a test tube containing 9 ml of physiological NaCl, incubated for 15 minutes. In a test tube containing flies, 1 ml of suspension is taken, then poured into a petri dish. Subsequently, the petri dish was added to the nutrient agar medium with a volume of 15 ml and then homogenized. The culture was incubated at 350 C for 2 x 24 hours. Each isolate that grew, was distinguished by colony shape, color, and edge of the colony in pure culture on two nutrient agar and incubated for 1 x 24 hours at 35°C. Colonies that grow in the subculture on sloping agar are then used for morphological analysis, gram staining, biochemical tests and DNA analysis.
Isolate bacteria from the fly’s digestive tract
Flies were preserved with 95% ethanol before being used as a source of bacterial inoculums, preserved again on 95% ethanol in new bottles for 60 minutes. After 60 minutes, flies are placed on sterile petri dishes with sterile filter paper to absorb the remaining ethanol on the surface of the fly’s body. Then, flies are noticed in the new sterile petri dish. Using a surgical instrument, cut longitudinally from the posterior direction (head) to the tail so that it is separated into two parts. Sterile cotton is applied from the head to the fly’s body parts, then inoculated in agar nutrient media. The culture was incubated at 35°C for 1 x 24 hours. Each isolate that grew, was distinguished by colony shape, color, and edge of the colony in pure culture on two nutrient agar and incubated for 1 x 24 hours at 35°C. gram, biochemical testing and DNA analysis.
Molecular identification
Preparation of Bacterial Isolate
Prior to DNA extraction and Purification, the purified bacterial isolate was previously inoculated into agar nutrient media and incubated for 1 x 24 hours at room temperature. Bacteria were isolated from the digestive tract of the green fly (Chrysomya megachepala) and the wing of the house fly (Musca domestica L.). Growing bacterial colonies are used as a source of bacterial cell suspension for DNA analysis.
Extraction and Purification of DNA
Bacterial suspension from pure culture is prepared aseptically. The bacterial isolate DNA was extracted with Presto TM Mini gDNA Bacteria Kit Geneaid as described in the manufacturer’s protocol, with modification by researchers. Suspension of bacterial isolates was taken as much (1 x 109 cells) / 200 µl, then put into a 1.5 ml sample tube. Subsequently added 200 µl of positive gram buffer (+), lysozyme (± 0.8 mg / 200 µl), then vortex until lysosim dissolves. Incubated at 37°C for 90 minutes. Next, add 20 µl proteinase K, then vortex. Incubate at 600C for 10 minutes, during incubation, the tube is rocked every 5 minutes. Next, add 200 µl GB of the buffer, mix with vortex for 10 seconds. Incubate at 70°C for 10 minutes, making sure the lisis is clear. Meanwhile, heat the elution on a thermostat with a temperature of 70°C. 200 µl of ethanol are added, which mixes quickly.
The GD column is inserted into the 2 ml collection tube, then transfer the mixture to the GD column. Centrifugation was carried out at 14000 -16000 g for 2 minutes. The collection tube is separated from the GD column, then enter the GD column in the new collection tube. 400 µl of W1 buffer was added to the GD column, then centrifuged at 16000 g for 30 seconds. Move the GD column to the new collection tube, then add 600 µl wash buffer to the GD column. Centrifuge at 16,000 g for 30 seconds, then move the GD column to the new collection tube. Centrifuge for 3 minutes at 16000 g to ensure dry/clean matrix column. Move the GD column to the new microcentrifuge tube. Add 100 µl of heated elution buffer and incubate for 3 minutes. Centrifuge at 16,000 g for 30 seconds. Transfer the total DNA to the DNA sample tube that has been previously labeled.
Analysis of DNA Concentration and Purity
The results of total DNA extraction were then analyzed for concentration and purity using a Lambda 35 Perkin Elmer UV / VIS Spectrometer. DNA purity can be seen with A260 / A280 ratio values between 1.8 – 2.0 nm. If <1.8 means that it is contaminated with protein and/or components of protein derivate contamination that affect DNA molecules, and if> 2.0 means contaminated with RNA (Protocol Kit), with the formula DNA Concentration and Purity21 as follows :
DNA concentration = absorbance 260 x 50
Purity of DNA = absorbance 260 / absorbance 280
Amplification of 16S rRNA gene by PCR method
Extracted DNA was amplified using the PCR method with Eppendorf Master cycles pro s PCR machine. The PCR process uses 2x MyTaq HS Red Mix Bioline and 16S rRNA primers used in the PCR amplification process were: 16sA (5´CGC CTG TTT AAC AAA AAC AT 3 ´) (Foward) and 16sB2 (5´TTT AAT CCA ACA TCG AGG 3´) (Reverse). The 16S RNA primer used can identify all types of eubacteria ( Cognato & Vogler, 2001). PCR conditions initial denaturation of 72°C for 50 seconds then denaturation following 94°C for 30 seconds. Annealing 49°C for 40 seconds, extension 72°C for 50 seconds, final extension 72°C for 5 minutes. The number of cycles as much as 35 times. (Table 1, Table 2 and Table 3). Visualization of PCR products using the agarose electrophoresis method of 1.8%. Electrophoresis results were observed using a UV Chamber to obtain electrogram images.
Table (1):
PCR Components.
PCR Component |
Volume (µL) |
|---|---|
2x MyTaq HS Red Mix Bioline |
25 |
Primer Forward 16sA |
1 |
Primer Reverse 16sB2 |
1 |
DNA of bacterial isolates |
2 |
DdH2O |
21 |
Total |
50 |
Table (2):
PCR Conditions According to the Protocol.
| Cyle | Duration (Seconds) | Temperature (°C) | Phase |
|---|---|---|---|
| 35 x | 50 | 72 | Initiating Denaturation |
| 30 | 94 | Denaturation | |
| 40 | 49 | Annealing | |
| 50 | 72 | Ekstension | |
| 300 | 72 | Final Ekstension |
Table (3):
Modified PCR Conditions.
| Cyle | Durasi (Detik) | Temperature (°C) | Fase |
|---|---|---|---|
| 35 x | 60 | 94 | Denaturation |
| 30 | 54 | Annealing | |
| 30 | 72 | Ekstension | |
| 60 | 72 | Final Ekstension |
Sequence processing and analysis
Sequencing uses ABI PRISM 3730xl Genetic Analyzer Developed by Applied Biosystems USA through Singapore’s FIRST BASE sequencing service. Sequencing results from Singapore’s First Base laboratory in the form of .seq file were analyzed using Geneious 10.1.3. The results of reading with Geneious 10.1.3 are sequence chromatograms, base sequences of 16S rRNA genes for each bacterial isolate and sequence characteristics. The consensus area was determined using Bioedit. Sequences of 16S rRNA gene consensus for each bacterial isolate obtained were used for homology analysis with BLAST (Basics Local Alignment Searching Tools) online on the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Selected BLAST result sequences were used to reconstruct phylogeny trees. Reconstruction of phylogeny trees using MEGA 7.0. The phylogeny tree model used was determined through the analysis of a suitable model in MEGA 7.0 software. The phylogeny tree construction uses two models with 1000x boostrap22. The phylogeny tree construction was also carried out online at the NCBI website.
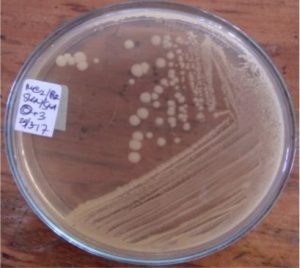
A total of seven bacterial isolates were successfully isolated and cultured pure. One isolate came from the digestive tract of a green fly and six isolates were successfully grown from the wing of a house fly (Table 4).
Table (4):
Bacterial isolates from the digestive tract of green flies and wing house flies in Luwuk city.
Isolate Code |
Origin of Isolates |
Pure Culture |
Staining |
|---|---|---|---|
S1 = USLHRS |
Digestion tract of green flies (Chrysomya megachepala L.) from hospital waste dumps. |
||
S2 = SKRMCPSR |
Isolated from the left wing of house flies (Musca domestica L.) from market waste dumps. |
||
S3 = SKNMCPSR |
Isolated from the right wing of house fly (Musca domestica L.) from the market garbage dump. |
||
S4 = SKNMCRS |
Isolated from the right wing of a house fly (Musca domestica L.) from a hospital waste dump. |
||
S5 = SKRMCPSR |
Isolated from the left wing of house flies (Musca domestica L.) from market waste dumps. |
||
S6 = SKRMCPSR |
Isolated from the left wing of house flies (Musca domestica L.) from market waste dumps. |
This isolate does not produce consistent gram staining. |
|
S7 = SKRMCPSR |
Isolated from the left wing of house flies (Musca domestica L.) from market waste dumps. |
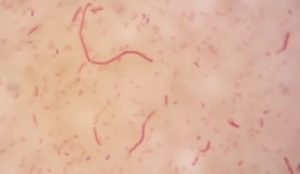
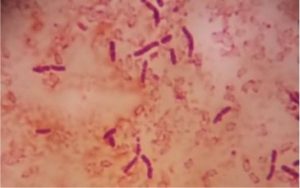
Five isolates had a white colony, one isolate had a grayish-white colony and one isolate had a yellow colony. As for the colony shape characteristics, two isolates had an irregular colony shape, three-round isolates, one isolate in the form of diffuse filaments and one isolate in the form of fine filaments. The characteristics of isolate cells were obtained by four gram-positive isolates, two gram-negative isolates and one unidentified isolate (Table 5).
Table (5):
Morphological characteristics of colonies of bacterial isolates from flies.
| NO. | Samples / Isolates Name | Morphology of bacterial colony | Gram Stain | |
|---|---|---|---|---|
| Colour | Shapes | |||
| 1 | S1 = USLHRS | White | Irregular | Gram positive(+) |
| 2 | S2 = SKRMCPSR | White | Irregular | Gram positive (+) |
| 3 | S3 = SKNMCPSR | White | Rounded | Gram positive (+) |
| 4 | S4 = SKNMCRS | White | Filament spread | Gram positive (+) |
| 5 | S5 = SKRMCPSR | Grayish white | Fine filament shape | Gram negative (-) |
| 6 | S6 = SKRMCPSR | White | Rounded | Not definition |
| 7 | S7 = SKRMCPSR | Yellom | Rounded | Gram negative (-) |
Extraction and Purification of total DNA of flies
DNA from a total of seven bacterial isolates was successfully extracted using Presto TM Mini gDNA Bacteria Kit Geneaid. Pure isolate culture is used as a cell source for the extraction and purification of total DNA. The total DNA volume of each isolate was 100 µl (approximately 75 ng). Based on the analysis of purity obtained the distribution of total DNA purity is 1.40 – 1.8. Whereas the distribution of total DNA concentration is 35.40 to 55.20 µg / µl (Table 6).
Table (6):
Concentration and purity of the extracted DNA.
No |
Samples / Isolates |
Concentration µg/ml (A260/A280) |
Purity (A260/A280) |
|---|---|---|---|
1 |
S1 = USLHRS |
41,20 |
1,80 |
2 |
S2 = SKRMCPSR |
47,35 |
1,75 |
3 |
S3 = SKNMCPSR |
38,20 |
1,43 |
4 |
S4 = SKNMCRS |
35,40 |
1,40 |
5 |
S5 = SKRMCPSR |
50,14 |
1,43 |
6 |
S6 = SKRMCPSR |
50,20 |
1,65 |
7 |
S1 = USLHRS |
50,10 |
1,70 |
Concentration and purity of total bacterial DNA greatly influence the success of 16S rRNA gene amplification by PCR method24,25. The optimal total DNA concentration based on the kit protocol used is 30 µg / µl up to 60 µg / µl, while for total DNA purity it is in the range of 1.7 – 2.0 (A260 / A280). Reach a conclusion, the concentration of DNA of seven bacterial fly isolates was good. The total DNA purity obtained is quite good. Modified protocols at the immersion stage of proteinase-K and RNAase have been shown to increase concentration but not to total DNA purity. However, the quality of the total bacterial DNA as a template will be known after the 16S rRNA gene amplification was performed using the PCR method.
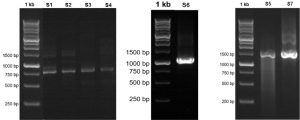
Fig. 1. Visualization of the 16S RNA gene amplicon of seven bacterial isolates from flies in Luwuk City, Central Sulawesi, Indonesia.
PCR and visualization of amplicon16S rRNA by electrophoresis method
Visualization of 16S rRNA gene amplicons of seven bacterial isolates in the range of 700 bp to 1500 bp. Isolates S1 was visualized at 780 bp, Isolate S2 800 bp, Isolate S3 815 bp, Isolate S4 890 bp, Isolate S5 1500 bp, Isolate S6 1097 bp, and Isolate S7 1500 bp. Based on the band formed, it indicates that the 16S rRNA gene amplification process was successful in all bacterial isolates (Fig. 2).
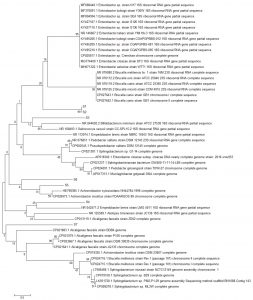
Fig. 2. Reconstruction of the phylogeny tree with neighbor joining model. The phylogeny tree construction uses two models with 1000x boostrap with MEGA 7.0.
Sequencing
The sequencing of 16S RNA gene nucleotides from seven bacterial fly isolates was carried out by the sequencing method. The sequencing product in the form of seq file, from Singapore’s FIRST BASE Laboratory, is read using the Geneous program. The consensus area of the 16S rRNA gene sequence was obtained using the Bioedit Program.
>Consensus Isolate S1
GCGCCTGTTTAACAAAAACAT ATGGCTTTGCAAAATCGAAAGATGAGG TATAAGGCCTGACACCTGCCCGGTGCTGG AAGGTTAAGAGGGGATGTCATCCGCAA GGAGAAGATTGAATCGAASCCCCAGTAAA CGGCKGCCGTAACTATAACGGTCCTAAKGYA GCAAATTCCTTGTCGGGAAGTTCCGACCTGC ACSAATGGTGTAACGATCTGGGCGCTGTCTCRC CRGAKCKCRGTSAAAKTGGGAKCGGTGAASAC GCCGATTACCCGAACRGGACRGAAASACC CCATGCACCTTCACTATRGCTTAACATTGAAATT GKGTACRRGATGTGTARGATAGGCGGGAGA TGTTGAAGTGGCTKCSCSAGGARTCTGGACA YYTGAAATAMYSCCMTWTCTGKAYYCRG TTWCYAAKTCRMCTGWGKCGAKRATGTTSST GGKYWGTWTKMCTGGGGTGGTCGCC TCCAAAAAGGTAACGGAGGCTTTCAAAG GTAAGCTCAGTACGCTTGGTAACCGTACGGG GAGTGCAATGGCATAAGCTTGCTTGACTGTGA GACCTACAAGTCGAGCAGGGTCGAAAGACGG ACATAGTGATCCGGTGGTTCTGTATGGAAG GGCCATCGCTCAAAGGATAAAAGGTACGCTGG GGA TAACAGGC
>Consensus Isolate S2
TTCTTT AGAAAAAG GGGGG GCCAGGAA GTTCGAATAAAAGGGGCAAGGTTTAACGCA CAGATTCATCGTGACGCCAATTTATTAGACTGAAG GGTTTCCCCTTGTCGGCCCCGGCTCCAAGACC TTCAGTCAACGCAAAGTGGCGAACCGTT GGCATGGG GCCCCGGAAAGCCCCAAAGAA CATGCTTGATGGTTCATTGACCGAGCTG CGCCCCTTCAAAGTGAGGACCTGAGTGA GGAGATCCTCAGCCCAAGGGCAT GATTGAGCAAGTGGCCGTGTTAATGGTAGCGCTGCC AGCAGAAAGTTCAGCATCTGAAGGACGTCGGCTCCAGCGTGAACTATCGCTTGATT GACGATCAGGGCCAGTC CCGCGAGGTGTGAATTACATGCTGCCTGTGGA GCTGGATGGCACCTGGTGTTTCTGGCTGGTA TGCGTTTGTAGTTCGGCGAGCCGTTACGGTA TGTGCGTTTCCCGGATGACGACAAGGGTTCCT GATAGAATTCATGGATTTGCGTGCGGCCAC GCAGAAAGAGGCCATGCTGCACCCAGGCGGCC GAGCGCTTTGCCGAGAGTAAGTCTTCGTC GCCCGAGCTAAAAGAGCTGAGACTCTCGG TATCCAGAGCCGTTTTGCAGACGTTTGCGCCAGCC GGTTTTG ATGGGTT GATCAGCGGTGTTC CAGAGGC TGAGCGA GAGAGTCT TCTGGG TCTGCAATTCCCAT GATTCAGTTCACGCCGTCGGAATTGCGTGATG CGATCAG GCCGCAACAAGCTTGGCCCGATCCAGAT CCCCGTCAGGAAAATAGCGCTCACACTCGCGGGATT GAGTCTGATGGACGATCATTCGCCAGCTCACGTGGGTGCCGGCCTTCGGCAG AGGCGGCTCTGTACA GCTGGGAGCAATATCAGTCCCGGGGGTA CAGGTTGA GTGCTGGCCTGGCATGGACGATCACCATCAGTCAGGTTCTCTACGTTGATGGGGCGCAGCTCGGT CTTCTGTATCGTCGATGCGTGTTCGTGGGTGCGGACTTGGCCTCCCATGCAAG TCAGCTCCATTTGCC ACATAGCGTAAAGTGTCGGAACCAGGGCTTTCCC AGGGATAGCCTTGCATGAAAGGGATAGTTCAGCCG TCGTAAA
>Consensus Isolate S3
TTTTTAATCTATCGCCTGTTCGCCTGTTTCCAGCAATATAGGGCTCTGCGAAGTCGCAAGTCGAC GTATAGGGTCTGACGCCTGCCCGGTGCTGGAAGGTTAAGAGGAGAGGTGCAA GCCTTGAATTSWAKCCCCAGTAAACGGCGGCCGTAACTATAACGGTCCTAASGTA SCGAAATTCCTTGTCKKGYAAGTTYCGACCTGCMCGAATGGCGTAACGAC TTCMCCGCTGTYTCCAACGCASACTCAGTGAAATTGAATTYCCCGTGAAGATGCGG GGTTCCTGCGGTTAGACGGAAAGACCCCGTGCACCTTTACTATAGCTTTACMCTG GCATTTGTGGCGACATGTGTAGGATAGGTGGTAGACTTTGAAGCGKAGGCG CCAGCCTTCGTGGAGTCATCCTTGAAATACCACCCTTATCGTCATAGATG TCTAACTGCGACCCGTATATCCGG GTCCAGGACCGTGTATGGTGGGTAGTTTGACTGGGG CGGTCGCCTCCTAAAGAGTAACGGAGGCGCGCRATGGTGGGCTCAGAACGGTCG GAAATCGTTCGTCGAGT GCAATGGCATAAGCCCGCCTGACTGCWAGACAGA CAAGTCGA GCAGAGACGAAAGTCGGTCATAGTGATCCRGTGGTCCCGTRTGGAAGGGCCATCGCTCAACGG ATAAAAGGTACGCCGGGGATAACAGGCTGATGACCCCCAAGAGTCCATATCGACG GAGTTGTTTGGCACCT CGATGTTGGATTAAAAGATTAATT
>Consensus Isolate S4
CCTGTTTTCGCCTGTTTTACAAAAACATA TGGCTTTGCAAAATCGAAAGATGAGGTATAAGGCCT GACACCTGCCCGGTGCTGGAAGGTTAAGAGGGAGATGTCATCGCAAGAG AAGCATTGAATCGAAGCCCCAGTAAACGGCGGCCGTAA CTATAACGGTCCTAAGGTAGC GAAATTCCTTGTCGGGTAAGTTCCGACCTGCACGAATGGTGTAACGATCTGGGCGCT GTYTCAGCCATGAGCT CGGTGAAATTGTGGTATCGGTGAAGACGCCGATTACCCGCA ACGGGACGGAAAGACCCCATGCACCTT CACTATAGCTTAACATTGAAATTGGGTACAGGATG TGTAGGATAGGCGGGAGATGTTGAAGTGG CTTCGCCAGGAGTCATGGAATCAACCTTGAA ATACCGCCCTTTCTGTATTCGGTTTCTAACTCG GCCATGCCGAGGACATTGTTTGGTGGGTAGTTT GACTGGGGTGGTCGCCTCCAAAAAGGTAACGGA GGCTTTCAAAGGTAAGCTCAGTACGCTTGGTAA CCGTACGTGGAGTGCAATGGCATAAGCTTGCTTGACTGTGAGACCAACAAGTCG ACCAGGGTCGAAAGA CGGACATAGTGATCCGGTGGTTCTGTATGGAAGGG CCATCGCTCAAAGGATAAAAGGTACGCTGGGGATAACAGGCTGATCTCCCCCAA GAGCTCATATCGACGGG GAGGTTTGGCACCTCTATGTTGGATTAAAATATAC
>Consensus Isolate S5
TAAGAGCTTGCTCTTATGAAGTTAGCG GCGGACGGGTGAGTAACACGTGGGTAACCTG CCCATAAGACTGGGATAACTCCGGGAAACCG GGGCTAATACCGGATAACATTTTGAACCGCATG GTTCGAAATTGAAAGGCGGCTTCGGCTGT CACTTATGGATGGACCCGCGTCGCATTAGC TAGTTGGTGAGGTAACGGCTCACCAAGGCAA CGATGCGTAGCCGACCTGAGAGGGTGATCGG CCACACTGGGACTGAGACACGGCCCAGACTCC TACGGGAGGCAGCAGTAGGGAATCTTCC GCAATGG ACGAAAGTCTGACGGAGCAACGCCGCGTGAGTGATGAAGGCTTTCGGGT CGTAAAACTCTGTT GTTAGGGAAGAACAAGTGCTAGTTGAATAAGCTGGCACCTTGACGG TACCTAACCAGAAAGCCACGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGTGG CAAGCGTTATCCGGAATTATTGGGC GTAAAGCGCGCGCAGGTGGTTTCTTAAGTCTGATG TGAAAGCCCACGGCTCAACCGTGGAGGGTCAT TGGAAACTGGGAGACTTGAGTGCAGAAGAG GAAAGTGGAATTCCATGTGTAGCGGTGAAATG CGTAGAGA TATGGAGGAACA CCAGTGGCGAAGGCGACTTTCTGGTCT GTAACTGACACTGAGGCGC
>Consensus Isolate S6
AGCTAGTATCCATCGTTTACGGCGTGGA CTACCAGGGTATCTAATCCTGTTTGCTCCCCACGCTTT CGTTCATCAGCGTCAATAAATACGTAGTAACCTGCCTT CGCAATTGGTATT CCATGTAATATCTAAGCATTTCACC GCTACACTACATATTCTAGTTACTTCCATACTACTCA AGCTAGACAGTATCAATGGCAGTGTCTT AGTTGAGCTAAGAAATTTCACCTCTGACTTAT CCTAACCGCCTACGAACCCTTTAAACCCAA TAATTCCGGATAACGCTCGCATCCTCCGTATTACCG CGGCTGCTGGCACGGAGTTAGCCGATCCTTATTC
>Consensus Isolate S7
AAGCGTTAATCGGAATTACTGGGC GTAAAGCG CACGCAGGCGGTC TGTCAAGTCGGATG TGAAATCCCCGGGCTCAACCTGGGAACTGCATTCGAAACTGGCAGGCTAGAGT CTTGTAGAGGGGGGT AGAATTCCAGGTGTAGCGGTGAAATGCGTAGA GATCTGGAGG AATACCGGTGGCGAAGGCGGCCC CCTGGACAAAGACTGACGCTCAGGTGCGAAA GCGTGGGGAGCAAACAGGATTAGATACCCT GGTAGTCCACGCCGTAAACGATGTCGACTTGG AGGTTGTGCCCTTGAGGCGTGGCTTCCGG AGCTAACGCGTTAAGTCGACCGCCTGGGGA GTACGGCCGCAAGGTTAAAACTCAAATGAATTGACGGGGGCCCGCACAAGC GGTGGAGCATGTGGT TTAATTCGATGCAACGCGAAGAACCTTACCTAC TCTTGACATCCAGAGAACTTTCCAGAGATGGATTGGTGCCTTCGGGAACTCTGAGACAGGTGCTGCA TGGCTGTCGTCAGCTCGTGTTGTGAAATGTTGGGTTAAGTCCCGCAACGAG CGCAACCCTTATCCTTTGTT GCCAGCGGTTCGGCCGGGAACTCAAAGGAGAC TGCCAGTGATAAACTGGAGGAAGGTGGGG ATGACGTCAAGTCATCATGGCCCTTACGAGTAG GGCTACACACGTGCTACAATGGCGCATACAAAG AGAAGCGACCTCGCGAGAGCAAGCGGACCTCA TAAAGTGCGTCGTAGTCCGGATTGGAGTCTG CAACTCGACTCCATGAAGTCGGAATCGCTAGTAAT CGTAGATCAGAATGCTACGGTGAATACGTTCCCGG GCCTTGTACACACC GCCCGTCACACCATGG
Each 16S RNA consensus gene sequence of each isolate was used for alignment using BLAST. Analysis of 16S RNA Isolate S1 gene sequence alignment showed a 90% similarity with Bifidobacterium minimum [NR 044692.2]. Isolate S2 showed 86% similarity with Alcaligenes faecalis [CP021883.1]. Isolate S3 had a similarity of 94% Brucella melitensis bv. [NR_076080.2]. Isolate S4 has a 99% similarity with Sphingobacterium sp. [LK931720.1]. Isolate S5 has a 100% similarity with Bacillus sp. [MH071158.1]. Isolate S6 has a 97% similarity with Gamma proteobacterium [KJ127178.1]. Whereas Isolate S7 has a similarity of 100% Enterobacter asburiae [MH071322.1] (Appendix 1).
House flies have long been known as vectors of various types of diseases caused by bacteria. The bacteria that are transmitted by house flies to humans are very dependent on habitat. Identification of bacteria by conventional methods not only requires a long time but also the accuracy of the species identified is still low. Molecular identification using the 16S rRNA gene has been widely used in determining the position of bacterial species. Bacterial identification based on morphology and biochemical analysis has been apply before on Musca domestica and Chrysomya megachepala that lives in Luwuk City, Central Sulawesi, Indonesia. Molecular identification can determine the position of bacterial species that exist in flies and construct phylogeny trees of these bacteria. Knowing the species of bacteria in flies is very important to overcome the transmission of pathogenic bacteria conducts by flies.
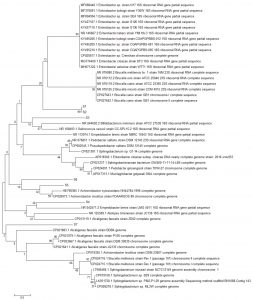
The phylogeny tree construction aims to determine the evolutionary history of a species, diversity characteristics, predict novelty of genes, break pathogenic microbial strains and other purposes20. The phylogeny construction is based on consideration of the type or model of the phylogeny tree, the sequence that becomes the comparison and calculation of sequence data21. In this present study, the term reconstruction is used because the phylogeny tree was rebuilt based on the 16S rRNA gene sequence of bacteria from flies with BLAST sequences that were recorded in the NCBI gene bank. The phylogeny reconstruction was executed using two models. It aims to compare and ensure phylogeny trees were formed. The phylogeny tree built using the Neighbor-Joining model has topography similar to the minimum evolution model (Fig. 3 in Fig. 4).
Fig. 3. Reconstruction of the phylogeny tree with minimum evolution model. The phylogeny tree construction uses two models with 1000x boostrap with MEGA 7.0.
Based on the 16S rRNA gene in constructing phylogenic trees, isolate S1 and S2 isolate have the closest evolutionary relationship with Brucella sp. Isolate S1 and Isolate S2 form the same node, thus having a very close evolutionary relationship. Isolate S3 has the closest evolutionary relationship with Bifidobacterium minimum even though it is in a different node. S4 isolate has the closest evolutionary relationship with Empedobacter brevis. Isolate S5 has the closest evolutionary relationship with Achromobacter xylosoxidans. Isolate S6 has the closest evolutionary relationship with isolate S5, then with Achromobacter xylosoxidans. Isolate S7 has the closest evolutionary relationship to Alcaligenes faecalis (Fig. 2 and Fig. 3).
From the results of this study, it is known that the similarity of 16S rRNA gene sequences is different from the close evolutionary relationship of each bacterial isolate obtained from green flies and house flies in Luwuk. The alignment of 16S rRNA sequences using BLAST (https://www.ncbi.nlm.nih.gov/) is based on the similarity in the sequence of nucleotide sequences of each isolate with similar sequences that have been recorded in the NCBI gene bank. In contrast to phylogeny reconstruction, it was built based on the close evolutionary relationship of each isolate based on 16S RNA sequences22,24-26. Species of bacteria identified from green flies and house flies that inhabit the city of Luwuk have never been reported to be associated with flies.
Research conducted by Park et al. 2019 states that the genus Staphylococcus and Weissella were predominantly found on the external portion of house flies however, in this study neither species of bacteria was found4. The genus Pseudomonas and E. coli were found the most on the surface of the body of the fly that lives in the hospital8. In this study, the bacteria isolated from flies that live in hospital waste bins were very different from those commonly found. Bacterial species found based on molecular identification are Bifidobacterium minimum (Isolate 1), Alcaligenes faecalis (Isolate 2), Brucella melitensis bv. (Isolate S3), Sphingobacterium sp. (Isolate S4), Bacillus sp. (Isolate 5), Gamma proteobacterium (Isolate S6), and Enterobacter asburiae (Isolate 7). Interestingly, the results of previous studies of the seven bacteria have antibiotic activity against pathogenic bacteria Escheresia coli and Staphylococcus sp1
Based on the alignment analysis of the BLAST method, the 16S RNA Isolate S1 gene sequence showed a 90% similarity with Bifidobacterium minimum [NR 044692.2]. Isolate S2 showed 86% similarity with Alcaligenes faecalis [CP021883.1]. Isolate S3 had a similarity 94% Brucella melitensis bv. [NR_076080.2]. Isolate S4 has a 99% similarity with Sphingobacterium sp. [LK931720.1]. Isolate S5 has a 100% similarity with Bacillus sp. [MH071158.1]. Isolate S6 has a 97% similarity with Gamma proteobacterium [KJ127178.1]. Whereas Isolate S7 has 100% similarity of Enterobacter asburiae [MH071322.1]. Based on the evolutionary relationship on phylogeny trees formed using 16 S RNA sequences and BLAST sequences, isolates S1 and isolates S2 have the closest evolutionary relationship with Brucella sp. Isolate S3 has the closest evolutionary relationship with Bifidobacterium minimum. Isolate S4 has the closest evolutionary relationship with Empedobacter brevis. Isolate S5 has the closest evolutionary relationship with Achromobacter xylosoxidans. Isolate S6 has the closest evolutionary relationship with isolate S5, then Achromobacter xylosoxidans. Isolate S7 has the closest evolutionary relationship to Alcaligenes faecalis. Bacteria found in house flies and green flies from the city of Luwuk were first reported to be associated with flies.
ACKNOWLEDGMENTS
We would like to acknowledge Prof. Dr. Ir. Max Tulung, MS for his assistance in the morphology identification of flies. Thank you to the chair and staff of the bioactivity and molecular biology laboratory, State University of Manado, for assisting in the identification of molecular bacteria.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This work was supported by the Ministry of Research, Technology and Higher Education through the Doctoral Research Dissertation Schema Scheme (Hdristekdikti 2018 / 2019-01)
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files.
- Kanan M, Tulung M. DNA barcoding for identification flies species from differents habitats in Luwuk, Sulawesi Tengah. J Entomol Zool Stud. 2018;6(2):2046-2052.
- Khamesipour F, Lankarani KB, Honarvar B, Kwenti TEA. systematic review of human pathogens carried by the housefly (Musca domestica L.). BMC Public Health. 2018;18(1):1049.
Crossref - Nazari M, Mehrabi T, Hosseini SM, Alikhani MY. Bacterial contamination of adult house flies (Musca domestica) and sensitivity of these bacteria to various antibiotics, captured from Hamadan City, Iran. Journal of Clinical And Diagnostic Research (JCDR). 2017;11(4):DC04.
Crossref - Park R, Dzialo MC, Spaepen S. Microbial communities of the house fly Musca domesticavary with geographical location and habitat. Microbiome. 2019;7:147.
Crossref - Bahrndorff S, de Jonge N, Skovgard H, Nielsen JL. Bacterial Communities Associated with Houseflies (Musca domestica L.) Sampled within and between Farms. PloS One. 2017;12(1):e0169753.
Crossref - Zhao X, Zhang X, Chen Z, Wang Z, Lu Y, Cheng D. The Divergence in Bacterial Components Associated with Bactrocera dorsalisacross Developmental Stages. Front Microbiol. 2018;9:114.
Crossref - Jacob JJ, Veeraraghavan B, Vasudevan K. Metagenomic next-generation sequencing in clinical microbiology. Indian J Med Microbiol. 2019;37:133-40.
Crossref - Vijayakumar S, Biswas I, Veeraraghavan B. Accurate identification of clinically important Acinetobacterspp.: an update. Future Sci OA. 2019;5(6):FSO395.
Crossref - Gupta AK, Nayduch D, Verma P, Shah B, Ghat HV, Patole S, Shouche YS. Phylogenetic characterization of bacteria in the gut of house flies (Musca domestica L.). FEMS Microbiol Ecol. 2012;79:581-593.
Crossref - Kassiri H, Akbarzadeh K, Ghaderi A. Isolation of pathogenic bacteria on the house fly, Musca domestica L.(Diptera: Muscidae), body surface in Ahwaz hospitals, Southwestern Iran. Asian Pacific Journal of Tropical Biomedicine. 2011;1116:S1119.
Crossref - Rotty IE, Pinontoan O, Tulung M, Rumengan I, Mokosuli YS. Molecular identification of house fly, Musca domestica L. (Diptera : Muscudae), using mitochondrial DNA partial genes cytochrome oxidase sub unit 1 (CO1) in Manado city. International J Entomol Research. 2018;3(2):168-176.
- Chaiwong T, Srivoramas T, Sueabsamran P, Sukontason K, Sanford, MR, Sukontason KL. The blow fly, Chrysomya megacephala, and the house fly, Musca domestica, as mechanical vectors of pathogenic bacteria in Northeast Thailand. Trop Biomed. 2014;31(2):336-46.
- Pava-Ripoli M, Pearson REG, Miller AK, Ziobro GC. Prevalence and relative ridk of Cronobacter spp., Salmonella spp., and listeria monocytogenes associated with the body surfaces and guts of individual filth flies. Appl Environ Microbiol. 2012;78:7891-7902.
Crossref - Ngoen-klan R, Moophayak K, Klong-klaew T, et al. Do climatic and physical factors affect populations of the blow fly Chrysomya megacephala and house fly Musca domestica? Parasitology Research. 2011;109:1279-1292.
Crossref - Macovei L, Miles B, Zurek L. The potential of house flies to contaminate ready-to-eat food with antibiotic resistant enterococci. J Food Protect. 2008;71:432-439.
Crossref - Chakrabarti S, Kambhampati S, Zurek L. Assessment of house fly dispersal between rural and urban habitats in Kansas, USA. J Kans Entomol Soc. 2010;83:172-188.
Crossref - Manandhar R, Gokhale S. Are Houseflies Still Important Vector of Gastrointestinal Infections?. J Bacteriol Parasitol. 2017;8:318.
Crossref - The new buzz on antibiotics. Availiable on : http://www.abc.net.au/science/articles/2002/10/01/689400.htm [Last accessed on 2019 Dec 07].
- Levchenko MA, Silivanova EA, Khusanovna R, Bikinyaeva, Balabanova GF. Efficacy of acetamiprid and fipronil fly baits against the housefly (Musca domesticaL.) under laboratory conditions. Veterinary world. 2018;11(7):953-958.
Crossref - Khamesipour F, Lankarani KB, Honarvar B, Kwenti TE. A systematic review of human pathogens carried by the housefly (Musca domestica L.). BMC Public Health. 2018;18(1):1049.
Crossref - Barbosa TM, Serra CR, La Ragione RM, Woodward MJ, Henriques AO. Screening for bacillus isolates in the broiler gastrointestinal tract. Appl Environ Microbiol. 2005;71(2):968-978.
Crossref - Tamura T, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0, Molecular Biology and Evolution, 2007; 24(8):1596-1599.
Crossref - Kumar, Sudhir, Koichiro Tamura, Masatoshi N. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5.2:150-163.
Crossref - Simandjuntak S, Mokosuli YS. Molecular Barcoding Based 16S rRNA Gene of Thermophilic Bacteria from Vulcanic Sites, Linow Lake, Tomohon. Materials Science Forum. 2018; 967;71-82.
Crossref - Simandjuntak S, Mokosuli YS. Isolation and Identification of Thermophilic Bacteria, Producer of Amylase Enzyme, from Lake Linow, North Sulawesi. J Pure Appl Microbiol. 2018;12(2):543-554.
Crossref - Manuahe C, Mokosuli YS, Roring VIY. Optimization of DNA extraction and the position of mosquito Species from southeast minahasa in North sulawesi using NADH dehydrogenase Gene and Cytochrome oxidase Sub Unit 1 Gene. J Entomol Zool Stud. 2016;4(4):498-508.
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.