ISSN: 0973-7510
E-ISSN: 2581-690X
Bacillus bacteria are advantageous antagonistic organisms that can be used as bio-control agents. This study is aimed at screening the antagonistic activity of different strains of isolated Bacillus bacteria and molecular identification of the superior chitinase producer strain against dermatophytes fungi. Soil samples were collected from different places of Kotoor city, Gharbia Governorate, Egypt and Al Madina Al Munawwarah, Kingdom of Saudi Arabia. A collection of Bacillus isolated from soil was tested in vitro against the dermatophytes: Microsporum sp. and Trichophyton sp. The bacterial strains Kh-B1 and Kh-B2 showed the highest antagonistic activity against dermatophytes pathogenic fungi. The highest amount of chitinase productivity (13.6 units/ml) was obtained from the original Bacillus strain (Kh-B1) at 3 days of incubation. BLAST analysis of the amplified 16S ribosomal RNA gene sequence identified the Bacillus strain (Kh-B1) as Paenibacillus macerans. Upon the mutation induction by UV light, the highest chitinase-producing mutant was Kh-UVB-4 as it showed 305.88 percent production higher than the wild-type strain. While, upon the mutation induction by EMS, the highest amount of chitinase produced was 54.8 units/ml by mutant Kh-ESB-20, and it has produced 402.94% more than the original untreated strain. The application of RAPD-PCR protocol using three 15-mer random primers was used to determine the genetic effects of mutagenic treatments on the wild type strain (Kh-B1) as well as to demonstrate the genetic variability between the five most chitinase producing mutants and the wild type (Paenibacillus macerans).
Bacillus strain, Chitinase producer, UV and EMS mutagens, antagonistic activity, RAPD-PCR
The Bacillus genus is a diverse group of Gram-positive bacteria, facultatively anaerobic, endospore-forming rod-shaped bacteria that involves both extremophiles and mesophiles.1 Although the generally accepted natural reservoir for these bacteria is soil, still they are usually spread into the environment.1 Metabolically these microorganisms are chemo-organotrophs and are reliant on organic compounds as sources of carbon and energy.2 Bacillus spp. is regarded as remarkable bio-control agent because of its ability to produce high resistant endospores which are usually resistant to different and acute environmental stress.3,4 By dint of their pervasiveness in nature, and metabolic and genetic diversity driving the production of several antibiotics and enzymes, Bacillus sp. has turned progressively alluring for various biotechnological applications such as processing of fermented foods and production of engineered industrial enzymes used in detergent and food industries.2 The capability to generate a broad range of antimicrobial compounds mostly active against fungi and bacteria is one of the principal characteristics common among Bacillus strains.1 Several studies have reported that Bacillus spp. restrict the growth of numerous plant pathogens by their antagonistic characteristics, with different modes of action like production of enzymes (chitinase and β 1,3-glucanase) and antibiotics (fengycin, surfactin, and iturin) that degenerate antifungal agents and fungal polymers as reported by.5-7
The chitinase enzyme is culpable for catalyzing the biological hydrolysis of chitin to its monomer N-acetyl-D-glucosamine. It is being produced by several types of microorganisms such as fungi, bacteria, yeast including others.8 Chitinase produced by fungi digests chitin and utilizes it as a carbon and energy source. Therefore, this extracellular hydrolytic enzyme is used as a controlling agent for the generation of fungal protoplasts. Therefore, chitinase has been used as a non-toxic alternative to chemical fungicides.8 In higher plants, the defense system in combination with fungal pathogens produces chitinases, while bacteria can produce chitinases to assimilate chitin as nitrogen and carbon source.9 The chitinolytic enzymes or chitinolytic microorganisms are used in the biocontrol of plant pathogenic fungi and insects as a target for biopesticides and have various potential biotechnological applications.10-13 The biological control of plant pathogens offers interesting substitute measures for agricultural diseases and pests management without adverse effects of the chemical fungicides. The chemical fungicides are generally expensive, that induce pollution for the environment, and may lead to resistance of pathogen as reported by.14,15 Desired bacterial strains can be developed by different methods as isolating organisms with high specificity, high capacity or by tailoring genetically improved organisms by mutagenesis as showed by.16-22 Several studies have reported that strain development can be used for producing several industrial enzymes like lipase, chitinase, cellulase, glucoamylase, and protease.23-27 The application of antagonists in the control and management of diseases incited by different pathogenic fungi is being extensively studied, but the diverse mechanism involved in the degradation of the fungus by bacteria is not well established.8
Therefore, the current study is aimed at in vitro screening of antagonistic activity of different strains of isolated Bacillus bacteria and molecular identification of the superior chitinase producer strain against dermatophytes fungi. We further assessed the generation of induced mutants by ultraviolet (UV) and ethyl methanesulfonate (EMS) mutagenesis from the wild-type Bacillus strain and evaluated its chitinase activity. We also compared various mutants to the wild-type strain through random amplified polymorphic deoxyribonucleic acid (RAPD) analysis using polymerase chain reaction (PCR).
The present research was done at Biology Department, Science Faculty (Girls), King Abdulaziz University (KAU). Jeddah, KSA in collaboration with Microbial Genetics and Genetics and Cytology Departments, National Research Centre (NRC), Cairo, Egypt.
Strains, media and growth conditions
On potato dextrose agar (PDA), Microsporum sp. and Trichophyton sp. fungi were cultivated for 10 days at 28°C (Merck, Germany). Bacillus strains produced chitinase using two fermentation media: fermentation medium 1 (FM1), a yeast nitrogen base (YNB) medium containing 0.2 percent (w/v) chitin, and fermentation medium 2 (FM2), a 250 ml Erlenmeyer flask containing 2 percent (w/v) chitin powder, 0.1 percent K2HPO4 and 0.05% MgSO47H2O at 37°C and pH 7.0 29, 30.
Soil samples, isolation and screening for antagonistic bacteria
Soil samples were collected in sterile small reagent culture tubes, labeled accurately, and maintained at 4°C until analysis in Kotoor, Gharbia Governorate, Egypt, and Al Madina Al Munawara, Saudi Arabia. By transferring 1 gm of a soil sample to 9 ml of sterile distilled water, vortexed swiftly, and serially diluted up to 10 -10 with sterile water, the bacteria were isolated. Then, 0.1 ml of each dilution was spread across the surface of FM1 plates and incubated at 37°C overnight. After 24 hours, plates were inspected for colony growth on the surface of FM1 after dilutions, and a few colonies with similar appearances were selected. To evaluate the antagonistic bacteria, the Trichophyton sp. and Microsporum sp. of fungi were cultured on the center plate containing PDA and incubated for 24 hours at 28°C. The tested Bacillus strains were inoculated around the fungal growth induced by Trichophyton sp. and Microsporum sp. at the edge of the petri dish plates and incubated for 72 hours at 28°C.
Molecular identification of bacteria
Using the GeneJET Genomic DNA Purification Kit (Thermo K0721, Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA) protocol, deoxyribonucleic acid (DNA) was isolated from the Bacillus strain (Kh-B1, Kotoor, Gharbia Governorate, Egypt). For PCR amplification of the 16S ribosomal gene, the Maxima Hot Start PCR Master Mix (Thermo K1051, Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA) was used, and the nucleotide sequences of the 16S primers used were forward primer-5′ AGAGTTTGATCCTGGCTCAG -3′ and reverse primer-5′-GGTTACCTTGTTACGACTT-3′. To clean up the PCR product, the GeneJET PCR Purification Kit was employed (Thermo K0701). The DNA from the PCR product was sequenced using forward and reverse primers on an Applied Biosystems (ABI) 3730xl DNA sequencer (GATC Biotech, Constance, Germany).
Phylogenetic analysis
Phylogenetic analysis Using the Basic Local Alignment Search Tool, the isolate16S ribosomal DNA (rDNA) sequence was compared to those in the National Center for Biotechnology Information (NCBI) GenBank database (BLAST). The sequences were compared to those of public databases’ reference taxa. The parameter model was used to determine evolutionary distances, and the neighbor-joining approach in MEGA6 software was used to build phylogenetic trees.31,32 The 16S rDNA sequence has been deposited as accession number MF150037 in the NCBI Gene Bank nucleotide sequence database.
Mutagenesis with Ultraviolet (UV)
Bacillus cells were suspended in water (sterile distilled) after a two-day growth period, and the suspension was exposed to UV light at a distance of 20 cm for 10 minutes. After that, the UV spore suspensions were protected from light for two hours. FM1 was diluted to the appropriate concentrations and incubated for two days at 37°C. For chitinase activity testing, the colonies that developed swiftly and in large numbers were put onto nutritional agar (NA) medium slants.
Mutagenesis with Ethyl methanesulfonate (EMS)
Bacillus cells from a two-day-old culture were suspended in sodium phosphate buffer (0.1M, pH 7.0) and treated for 60 minutes with 200 mM EMS. FM1 was treated with appropriate dilutions and incubated for two days at 37°C. For the forward test activities of chitinase, the rapidly growing colonies were transplanted to NA medium slants
Chitinase activity measurement
Chitin was used as a colloidal substrate to test chitinase activity. One ml of 1.5 percent (w/v) colloidal chitin suspension of phosphate buffer (50 mM, pH 7.0) was combined with 0.5 ml of enzyme solution and placed for 15 minutes at 37°C. Using the Imoto and Yagishita approach33 and N-acetylglucosamine as the reference substrate, the amount of reducing sugar generated in the supernatant after centrifugation at 8000 rpm for 5 minutes (Sigma, Germany) was measured. One unit of chitinase activity was defined as the amount of enzyme that produced 1 µmol of reducing sugar per minute.
Genomic DNA extraction from mutant strains
The i-genomic BYF DNA extraction Mini Kit was used to isolate total DNA (iNtRON Biotechnology Inc., South Korea). The UV-absorbance at 260 nm and 280 nm of the acquired DNA was calculated using Shimadzu UV-VIS model UV-240 spectrophotometer.34 Molecular profiles of genetically excellent mutants The procedures outlined by35 were used to create mutant molecular DNA banding profiles. PCR amplification was carried out using PCR Beads (Life Sciences, UK). With the exception of the DNA
Molecular profiles of genetically excellent mutants
The procedures outlined by35 were used to create mutant molecular DNA banding profiles. PCR amplification was carried out using PCR Beads (Life Sciences, UK). With the exception of the DNA template and primer, each bead includes all of the ingredients needed to run a 25 µl PCR amplification experiment. Operon Technologies Inc, Venlo, Netherlands, provided three distinct primers for this investigation. The primer sequences are as follows: first primer (P1)- 5′- CATACCCCCGCCGTT-3′, second primer (P2)-5′- GTGTTGTGGTCCACT-3′, and third primer (P3)-5′- TGAGTGGTCTACGTG-3′. Each PCR bead received a random primer (12 ng) and a purified DNA sample (40 ng). Using sterile distilled water, the total amount of the reaction cocktail was up to 25 µl.
The amplification was carried out in accordance with the following protocol:
Step 1: Denaturation of double-stranded DNA for 5 minutes at 95°C.
Step 2: Anneal the primer to the template DNA for 2 minutes in accordance with primer GC-ratio.
Step 3: Polymerization of DNA by incubating at 72°C for 2 minutes.
The processes above were then recycled 35 times. Gel electrophoresis was used to separate the DNA products obtained from the RAPD investigation by size. These products were loaded into precast wells on a 1.0 percent agarose gel with 1 X Tris/Borate/EDTA (TBE) buffer and ran for around 2 hours at a constant current of 100 volts. A 100-base pair (bp) ladder (Vivantis-Technologies # NL 1407- Malaysia) was used to identify the various bands. After staining with ethidium bromide, the bands were photographed using a Gel Documentation System with a UV transilluminator.
Statistical analysis
SPSS (Statistical Package for Social Science) Version 16.0 was used for statistical analysis (SPSS Inc., Chicago, IL, USA).
The information was presented in the form of a number and a percentage.
Screening of the collected strains against some dermatophytes fungi
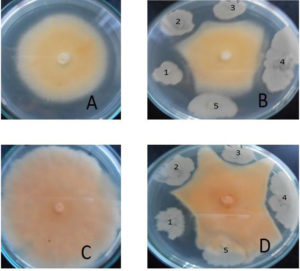
For the screening of antifungal activity of selected strains against some dermatophytes fungi (Trichophyton sp. and Microsporum sp.), the five isolated bacterial strains from several isolated bacterial isolates were selected on FM1 based on their growth rate (colony growth area) and tested against the selected dermatophytes fungi. The results showed that most bacterial strains have different appearances of antagonistic activities against the dermatophytes fungi (Fig. 1). The bacterial strain (Kh-B1) has shown the remarkable antagonistic activity effects against all tested dermatophytes pathogenic fungi. The Bacillus is widespread in the environment and is found in the soil, dust, air, and water, and is effective antagonist of many pathogens.36 Several studies have confirmed that Bacilli have efficient antimicrobial activity among the soil bacteria,37-39 Inconsistently with our results, a study from India showed the antimicrobial potential of Bacillus subtilis.40 Also, a study carried out in Thailand showed that Bacillus pumilus isolated from Thailand soil produced a new product of antimicrobial peptide.36 To further support our results, the antifungal activity of isolated Bacillus sp. was shown by.41,42 Furthermore, another study described those cytosolic proteins of Escherichia coli due to their antifungal potential as active against pathogenic strains of Candida albicans, Aspergillus niger, Aspergillus fumigatus, and Aspergillus flavus.38,39 In one more study, it was concluded that Escherichia coli showed absolute suppression of pathogenic fungi like Aspergillus niger and Aspergillus flavus.43
Fig. 1. Antagonistic effects of bacterial strains isolated from soil against dermatophytic fungi.
The plates A (control) and B shows antagonistic effects on Microsporum sp. The plates C(control) and D shows antagonistic effects on Trichophyton sp. The bacterial strains used are in the following sequence (1) Kh-B1, (2) Kh-B2, (3) Kh-B3, (4) M36 and (5) M14.
Assessment of the selected strains and production media
Table 1 shows the influence of two production media (FM1 and FM2), as well as incubation durations (2, 3 and 5 days), on the chitinase production of the collected Bacillus strains. The findings in Table 1 clearly demonstrate that FM1 was the best, since all of the tested Bacillus strains generated more chitinase on this medium after varied incubation durations. Over 3 days of incubation, the original Bacillus strain (Kh-B1) produced the most chitinase production (13.6units/ml). In terms of chitinase production on the FM1, the Bacillus strain (Kh-B2) trailed Kh-B1. The lowest quantity of chitinase synthesis, on the other hand, was 10.3 units/ml generated by Bacillus strain (M14) on FM2 after 2 days of incubation. In comparison to the other tested strains, Bacillus (Kh-B1) produced most of the chitinase on FM1 of the five tested strains.
Table (1):
Shows the effects of varied fermentation media and incubation duration on the selected Bacillus strains chitinase production grown in shaking flasks as batch fermentation.
| Bacillus strains Code | Chitinase activity (in units/ml) in fermentation medium 1 (FM1) | Chitinase activity (in units/ml) in fermentation medium 2 (FM2) | ||||
|---|---|---|---|---|---|---|
| Incubation time (day) | Incubation time (day) | |||||
| 2 d | 3 d | 5 d | 2 d | 3 d | 5 d | |
| Kh-B1 | 12.3 | 13.6 | 13.5 | 11.6 | 13.2 | 13.0 |
| Kh-B2 | 12.1 | 12.3 | 12.3 | 11.2 | 11.7 | 11.6 |
| Kh-B3 | 11.8 | 12.0 | 12.1 | 11.3 | 11.4 | 11.4 |
| M36 | 11.4 | 11.7 | 11.9 | 10.5 | 10.9 | 10.7 |
| M14 | 11.1 | 11.3 | 11.5 | 10.3 | 10.6 | 10.5 |
As a result, Bacillus (Kh-B1) in this work was used as the initial strain for all of the (genetic methods) used for chitinase production enhancement on FM1. The high production of chitinase enzyme show significant control against the dermatophyte’s fungi. These observations are inconsistent with the study where Bacillus subtilis exhibits the enhanced production of chitinase.30 The chitinase enzyme can degrade chitin, and therefore can be easily used against phytopathogenic fungi as a biological fungicide. The bacterial chitinase enzymes have been used to protect against the parasites in fungi, especially in the breakdown of micelles of fungal pathogens. A study did also show that Bacillus subtilis produced a high amount of chitinase enzyme with antifungal properties and therefore is an excellent choice in the application of biotechnology, food, and pharmacology industries.44
Identification of Bacillus (Kh-B1)
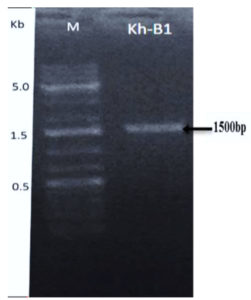
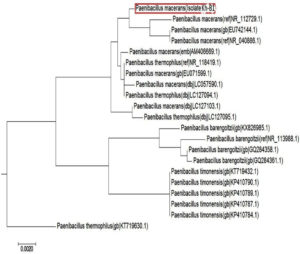
The 16S primers (forward and reverse) were used to amplify the area of the 16S ribosomal ribonucleic acid (rRNA) gene from Bacillus genomic DNA (Kh-B1). Following PCR amplification, a band of about 1500 bp was produced (Fig. 2). The amplified 16S rRNA gene sequence was blasted and found to be 99 percent identical to the partial 16S rRNA gene of the Paenibacillus macerans NBRC15307 strain (accession number: NR112729.1). The Bacillus strain (Kh-B1) was identified as Paenibacillus macerans based on this resemblance (Fig. 3). Senol et al., 2014 discovered that the strain of Paenibacillus macerans has antifungal properties as well as high chitinase synthesis.44
Fig. 2. Agarose gel image showing the amplified band of 16S-DNA for bacterial strain (Kh-B1) using 16S primers.
The ladder DNA marker used has three remarkable bands 500 bp or 0.5 kb, 1500bp or 1.5kb, 5000bp or 5kb (lane M).
Fig. 3. A phylogenetic tree of the effective Bacillus strain (Kh-B1) that produces chitinase in comparison to the most closely related bacterial strain inside the database (Paenibacillus macerans).
Mutation induction and chitinase activity
Table 2 compares the chitinase production of various Bacillus (Kh-B1) exposed to UV-light for 10 minutes to the untreated parent strain. From Kh-UVB-1 through Kh-UVB-29, these UV-mutants were given certain names. These findings revealed that the most of the examined mutants generated more chitinase than their original parental strain. However, 13 mutants were shown to be somewhat less effective than the original strain in terms of chitinase activity. A Kh-UVB-4 was the most productive chitinase mutant, generating 305.88 percent more than the normal strain. In terms of chitinase output, the mutant Kh-UVB-23 trailed Kh-UVB-4, producing 230.15 percent more than the normal strain. Table 3 compares the chitinase productivity of various mutants generated by exposing the parental strain to EMS-mutagen for 60 minutes to that of the parental strain. Following the same procedure, these EMS- mutants were designated Kh-ESB-1…Kh-ESB-29.
Table (2):
Productivity of chitinase for different mutants obtained by the exposure of Bacillus (Kh-B1) to UV-light for 10 minutes.
Bacillus strains Code |
Chitinase output (units per ml) |
% Greater than the original strain |
Bacillus Strain Code |
Chitinase output (units per ml) |
% Greater than the original strain |
|---|---|---|---|---|---|
Wild type (Kh-B1) |
13.6 |
100.00 |
Kh-UVB -15 |
13.5 |
99.26 |
Kh-UVB-1 |
29.4 |
216.18 |
Kh-UVB -16 |
28.3 |
208.09 |
Kh-UVB -2 |
17.0 |
125.00 |
Kh-UVB -17 |
12.8 |
94.12 |
Kh-UVB -3 |
12.5 |
91.91 |
Kh-UVB -18 |
24.5 |
180.15 |
Kh-UVB -4 |
41.6 |
305.88 |
Kh-UVB -19 |
12.8 |
94.12 |
Kh-UVB -5 |
11.6 |
85.29 |
Kh-UVB -20 |
31.0 |
227.94 |
Kh-UVB -6 |
13.5 |
99.26 |
Kh-UVB -21 |
24.5 |
180.15 |
Kh-UVB -7 |
13.2 |
97.05 |
Kh-UVB -22 |
10.8 |
79.41 |
Kh-UVB -8 |
14.5 |
106.62 |
Kh-UVB -23 |
31.3 |
230.15 |
Kh-UVB -9 |
12.9 |
94.85 |
Kh-UVB -24 |
18.7 |
137.50 |
Kh-UVB -10 |
19.2 |
141.18 |
Kh-UVB -25 |
19.5 |
143.38 |
Kh-UVB -11 |
15.5 |
113.97 |
Kh-UVB -26 |
10.5 |
77.21 |
Kh-UVB -12 |
23.4 |
172.06 |
Kh-UVB -27 |
11.6 |
85.29 |
Kh-UVB -13 |
13.3 |
97.79 |
Kh-UVB -28 |
29.4 |
216.18 |
Kh-UVB -14 |
29.9 |
219.85 |
Kh-UVB -29 |
12.9 |
94.85 |
The results obtained from the EMS-mutants showed an opposite trend to those reported from the UV-mutants because the effect of a mutagen varies depending on the type of mutagen, organism, condition, stage of growth, and other factors, and the majority (20 out of 29) of the tested EMS-mutants produced chitinase at a higher level than their original parental strain. The mutant Kh-ESB-20 generated most of the chitinase, 54.8units/ml, and it produced 402.94 percent more than the original untreated strain. Following the same procedure, 19 mutants generated chitinase that was greater than the original strain but lower than the good mutant Kh-ESB-20. The mutant Kh-ESB-12 produced 393.38 percent more chitinase than the original strain, thus following the Kh-ESB-20 in chitinase productivity. The above findings are in agreement with some of the previous studies done to enhance the production of chitinase by mutations. A study by Aly et al. showed the chitinase production has been improved by Bacillus pumilus CHT11 using UV-mutagenesis.20
Furthermore, UV treatments have been shown to generate mutants with increased chitinase synthesis as well as bio-control activity against several pathogenic fungi.20 It has also been observed that the mutant Bacillus pumilus (BM33) produced five times more chitinase than the wild-type strain.20 Research found that treating Bacillus subtilis with the chemical mutagen EMS increases the synthesis of three mucolytic enzymes (β-1,3-glucanase, cellulase, and chitinase).21 The superior mutant Bacillus subtilis M59 produced more mucolytic enzyme and inhibited the development of plant fungal pathogens through enzyme-mediated inhibition than the wild-type strain and other Bacillus subtilis mutants.21 Another study found that N-methyl-N’-nitro-N-nitrosoguanidine (NTG) treatment or UV radiation resulted in strain formation for hyperproduction of extracellular chitinase enzyme in Pseudomonas stutzeri.16 Also, following mutagenesis, good mutants (Pseudomonas stutzeri YPL-M26 and YPL-M178) produced bigger inhibitory zones.
Table (3):
The productivity of chitinase for several mutants obtained by the treatment of Bacillus (Kh-B1) with EMS for 60 minutes.
Bacillus strain s Code |
Chitinase output (units per ml) |
% Greater than the original strain |
Bacillus Strain Code |
Chitinase output (units per ml) |
% Greater than the original strain |
|---|---|---|---|---|---|
Wild type (Kh-B1) |
13.6 |
100.00 |
Kh-ESB-15 |
31.0 |
227.94 |
Kh-ESB-1 |
25.8 |
189.71 |
Kh-ESB-16 |
35.5 |
261.03 |
Kh-ESB-2 |
24.5 |
180.15 |
Kh-ESB-17 |
12.5 |
91.91 |
Kh-ESB-3 |
17.6 |
129.41 |
Kh-ESB-18 |
41.5 |
305.15 |
Kh-ESB-4 |
11.6 |
85.29 |
Kh-ESB-19 |
29.9 |
219.85 |
Kh-ESB-5 |
15.7 |
115.44 |
Kh-ESB-20 |
54.8 |
402.94 |
Kh-ESB-6 |
10.5 |
77.21 |
Kh-ESB-21 |
15.5 |
113.97 |
Kh-ESB-7 |
27.6 |
202.94 |
Kh-ESB-22 |
13.0 |
95.59 |
Kh-ESB-8 |
24.5 |
180.15 |
Kh-ESB-23 |
13.5 |
99.26 |
Kh-ESB-9 |
28.3 |
208.09 |
Kh-ESB-24 |
12.5 |
91.91 |
Kh-ESB-10 |
33.8 |
248.53 |
Kh-ESB-25 |
50.5 |
371.32 |
Kh-ESB-11 |
42.4 |
311.76 |
Kh-ESB-26 |
29.5 |
216.91 |
Kh-ESB-12 |
53.5 |
393.38 |
Kh-ESB-27 |
12.8 |
94.12 |
Kh-ESB-13 |
10.5 |
77.21 |
Kh-ESB-28 |
42.5 |
312.50 |
Kh-ESB-14 |
11.6 |
85.29 |
Kh-ESB-29 |
13.3 |
97.79 |
With UV or NTG have been obtained. Due to the synthesis of chitinase and laminarinase, these mutants are thought to have improved antifungal activity. It has also been reported that the antifungal mechanism of Pseudomonas stutzeriYPL-1 rests more on enzymatic lysis of the cell wall components of Fusarium solani by chitinase than on laminarinase.16 Furthermore, a study has confirmed that mutants of Pseudomonas alcaligenes strain treated by UV for 10 minutes produced a maximum amount (119.55 ± 4.29 units/ml) of chitinase, while the wild strain produced only 79.2 ± 1.41 units/ml of chitinase.22
Molecular analysis of mutants
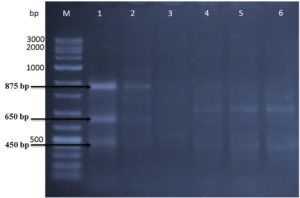
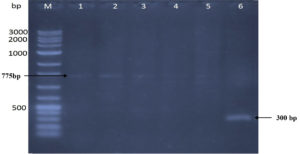
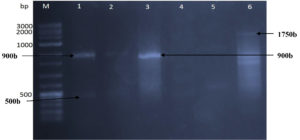
RAPD analysis utilizing three 15-mer random primers was used to assess the genetic influence of mutagenesis treatments on the nucleotide sequence of the five DNA mutants with the greatest chitinase production relative to the original strain (Kh-B1). Using primer-1, the findings revealed obvious changes in band number and size between the original strain and its mutations (Fig. 4). Three amplified sharp bands with sizes 450 bp, 650bp, and 875 bp were clearly found (lane 1) when the template used was the wild type strain DNA. The same amplified band sizes of wild-type strain were found when the Kh-UVB-4 DNA was utilized (lane 2), but the bands were faint and an additional two more faint bands were also detected with sizes 800 bp and 1000bp. On the other hand, each of the three mutants Kh-ESB-12, Kh-ESB-20, and Kh- ESB-25 exhibited three faint amplified bands with sizes 450 bp, 500 bp and 750 bp (lanes 4, 5 and 6). Another six very faint bands with sizes of 450 bp, 500 bp, 650 bp, 800 bp, 875 bp and 1000 bp were observed when the Kh-UVB-23 DNA was utilized (lane 3). The primer-2 profiles of the original strain and the mutations studied are shown in Fig. 5. Two bands with sizes of 750 bp and 775 bp were detected (lane 1). Additionally, all mutants (lanes 2, 3, 4, 5 and 6) showed amplified banding profiles same as that of wild-type strains but the mutant Kh-ESB-25 (lane 6) exhibited an additional band of 300 bp. On using primer-3, the results showed apparent differences in band numbers and sizes between the original strain and its mutants (Fig. 6).
Fig. 4. The gel picture showed amplified banding patterns of DNA for 5 efficient mutants in relation to the original strain determined by RAPD.
Original strain using primer-1 is loaded in lane 1 against VC100 bp plus DNA ladder (Vivantis-technologies # NL 1407, Malaysia) loaded in lane M. Mutants were loaded in the following sequence: (lane 2) Kh-UVB-4, (lane 3) Kh-UVB-23, (lane 4) Kh-ESB-12, (lane 5) Kh-ESB-20 and (lane 6) Kh-ESB-25.
Fig. 5. The gel picture showed amplified banding patterns of DNA for 5 efficient mutants in relation to the original strain determined by RAPD. Original strain using primer-2 is loaded in lane 1 against VC100 bp plus DNA ladder (Vivantis-technologies # NL 1407, Malaysia) loaded in lane M. Mutants were loaded in the following sequence: (lane 2) Kh-UVB-4, (lane 3) Kh-UVB-23, (lane 4) Kh-ESB-12, (lane 5) Kh-ESB-20 and (lane 6) Kh-ESB-25.
Fig. 6. The gel picture showed amplified banding patterns of DNA for 5 efficient mutants in relation to the original strain determined by RAPD.
Original strain using primer-3 is loaded in lane 1 against VC100 bp plus DNA ladder (Vivantis-technologies # NL 1407, Malaysia) loaded in lane M. Mutants were loaded in the following sequence: (lane 2) Kh-UVB-4, (lane 3) Kh-UVB-23, (lane 4) Kh-ESB-12, (lane 5) Kh-ESB-20 and (lane 6) Kh-ESB-25.
Two amplified bands of size 500 bp and 900 bp were detected while the wild-type strain DNA was used as a template (lane 1). Another two amplified bands of the same size as wild type strain were seen when the Kh-UVB-4 DNA was utilized (lane 2). A single sharp band of 900 bp was detected when the Kh-UVB-23 DNA was utilized (lane 3). Two very faint amplified bands of 500 bp and 650 bp were observed when the Kh-ESB-12 DNA was used as a template (lane 4). Moreover, another two very faint amplified bands of 600 bp and 950 bp were seen when the Kh-ESB-20 DNA was utilized (lane 5). Five amplified bands with sizes of 600 bp, 700 bp, 800 bp, 875 bp, and 1750 bp were found when the Kh-ESB-25 DNA was used as a template (lane 6).
The above results showed complete conformity with the previous studies which have demonstrated the genetic effects of mutagenic treatments by EMS or UV on mutants of Bacillus megaterium and Bacillus amyloliquefaciens in comparison to their original strains. The results of these studies showed the same trend as found in our study such as differences in band numbers and sizes between both the original strain and its mutants, analyzed by RAPD.45,46 Moreover, in one more study, many changes have been detected in mutants compared to the parental strain.47 These changes in RAPD patterns revealed the verification of genetic changes of the DNA in mutants after the application of UV-light to induce mutation. It has also been demonstrated that the use of the RAPD procedure on some superior mutants leads to connecting the DNA changes of the mutants with significant capacity of biosorption potential.47 Furthermore, the study by Gomaa et al. in 2018 demonstrated that the biocontrol efficacy was statistically far better in the mutants in relation to wild-type strains. Furthermore, they employed RAPD and ISSR PCR methods to discover differences in DNA profiles between mutant and wild-type strains in addition to gamma-radiation treatments.48 Thus, from the above result and discussion, we can say that our study identified a strain of Bacillus (Paenibacillus macerans) with high antagonistic activity against dermatophytes fungi and a novel mutant with high chitinase activity. The characterization of these superior mutants by RAPD fingerprint confirmed the genetic changes in the mutant with high chitinase activity. The above-mentioned findings show that mutagenesis is a viable method to enhance chitinase activity for agricultural, industrial, and other objectives, and that it may also be employed against fungus.
In conclusion, the isolates of Bacillus strain (Kh-B1) proved very essential in obtaining the superior chitinase producer strains. The Bacillus strain (Kh-B1), the larger chitinase producer strain was identified as Paenibacillus macerans. The improvement was observed in chitinase activity by Paenibacillus macerans mutants after UV and EMS mutagenesis. Moreover, the use of RAPD-PCR application of UV and EMS mutagens to induce mutation led to the detection of some changes in RAPD profiles which were used to demonstrate the genetic variability of Paenibacillus macerans mutants.
ACKNOWLEDGMENTS
The authors are grateful to the Biology Department, Faculty of Science, University of King Abdulaziz, Jeddah, KSA, for providing us with laboratory space and additional support in order to conduct this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Baruzzi F, Quintieri L, Morea M, Caputo L. Antimicrobial compounds produced by Bacillus spp. and applications in food. Science Against Microbial Pathogens: Communicating Current Research and Technological Advances. 2011;2(1):1102-1111.
- Graumann PL. Bacillus. Caister Academic Press. 2017.
- Wang Z, Jiang M, Chen K, et al. Biocontrol of Penicillium digitatum on postharvest citrus fruits by Pseudomonas fluorescens. J Food Qual. 2018;2018:2910481.
Crossref - Romero D, de Vicente A, Rakotoaly RH, et al. The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaerafusca. Mol Plant Microbe Interact. 2007;20(4):430-440.
Crossref - Jiang YM, Zhu XR, Li YB. Postharvest control of litchi fruit rot by Bacillus subtilis. LWT-Food Science and Technology. 2001;34(7):430-436.
Crossref - Pinchuk IV, Bressollier P, Sorokulova IB, Verneuil B, Urdaci MC. Amicoumacin antibiotic production and genetic diversity of Bacillus subtilis strains isolated from different habitats. Res Microbiol. 2002;153(5):269-276.
Crossref - Leelasuphakul W, Hemmanee P, Chuenchitt S. Growth inhibitory properties of Bacillus subtilis strains and their metabolites against the green mold pathogen (Penicillium digitatum Sacc.) of citrus fruit. Postharvest Biol Technol. 2008;48(1):113-121.
Crossref - Kannan N, Shivam K, Balaji S, Rajashekar V, Murty RV. Optimization of chitinase production from Serratia marcescens-A classical approach. Biological Segment. 2010;1(1):1-23.
- Chang WT, Chen YC, Jao CL. Antifungal activity and enhancement of plant growth Bacillus cereus grown on chitin wastes. Bioresour Technol. 2007;98(6):1224-1230.
- Gupta V, Schmoll M, Herrera-Estrella A, Upadhyay R, Druzhinina I, Tuohy M (Eds.). Biotechnology and biology of Trichoderma.Newnes. 2014.
- Muzzarelli RA. Chitins and chitosans as immunoadjuvants and non-allergenic drug carriers.Marine Drugs 2010;8(2):292-312.
Crossref - Patil RS, Ghormade V, Deshpande MV. Chitinolytic enzymes: an exploration. Enzyme Microb Technol. 2000;26(7):473-483.
Crossref - Halder SK, Pal S, Mondal KC. Biosynthesis of fungal chitinolytic enzymes and their potent biotechnological appliances. In Recent Advancement in White Biotechnology Through Fungi. Springer. 2019:281-298.
Crossref - Kishore GK, Pande S. Chitin-supplemented foliar application of chitinolytic Bacillus cereus reduces severity of Botrytis gray mold disease in chickpea under controlled conditions. Lett Appl Microbiol. 2007;44(1):98-105.
Crossref - Reyes-Ramirez A, Escudero-Abarca BI, Aguilar-Uscanga G, Hayward-Jones PM, Barboza-Corona JE. Antifungal activity of Bacillus thuringiensis chitinase and its potential for the biocontrol of phytopathogenic fungi in soybean seeds. J Food Sci. 2004;69(5):M131-M134.
Crossref - Lim HS, Kim YS, Kim SD. Pseudomonas stutzeri YPL-1 genetic transformation and antifungal mechanism against Fusarium solani, an agent of plant root rot. Appl Environ Microbiol. 1991;57(2):510-516.
Crossref - Bae W, Mulchandani A, Chen W. Cell surface display of synthetic phytochelatins using ice nucleation protein for enhanced heavy metal bioaccumulation. J Inorg Biochem. 2002;88(2):223-227.
Crossref - Li PS, Tao HC. Cell surface engineering of microorganisms towards adsorption of heavy metals. Crit Rev Microbiol. 2015;41(2):140-149.
Crossref - Huang CC, Su CC, Hsieh JL, Tseng CP, Lin PJ, Chang JS. Polypeptides for heavy-metal biosorption: capacity and specificity of two heterogeneous MerP proteins. Enzyme Microb Technol. 2003;33(4):379-385.
Crossref - Aly MM, Tork S, Alakilli SY. Molecular characterization of chitiniolytic Bacillus pumilus isolated from marine habitats and enhancement of chitinase production by mutation. Univ Scholar in Biotechnol. 2011;1:14-21.
- Narasimhan A, Suresh S, Bist D, Shivakumar S. Enhancement of mycolytic activity of an antagonistic Bacillus subtilis through ethyl methane sulfonate (EMS) mutagenesis. Turk J Biol. 2013;37(3):323-328.
Crossref - Suji HA, Palevesam TA, Immanuel G, Raj S. Effect of different growth parameters on chitinase enzyme activity. African Journal of Biotechnology. 2014;13(23).
- Kucerova H, Vachova L, Chaloupka J. Mutants of Bacillus megaterium with altered synthesis of an exocellular neutral proteinase. Folia Microbiologica. 1984;29(2):99-103.
Crossref - Haki GD, Rakshit SK. Developments in industrially important thermostable enzymes: a review. Bioresour Technol. 2003;89(1):17-34.
Crossref - Bapiraju KVVSN, Sujatha P, Ellaiah P, Ramana T. Mutation induced enhanced biosynthesis of lipase. African Journal of Biotechnology. 2004;3(11):618-621.
- Al-Turki AI, Khattab AA, Ihab AM. Improvement of glucoamylase production by Aspergillus awamori using microbial biotechnology techniques. Biotechnology. 2008;7(3):456-462.
Crossref - Khattab AA, Abdel-Aziz MS. Mutation induction and protoplast fusion of Streptomyces spp. for enhanced alkaline protease production. J Appl Sci Res. 2012;8:807-814.
- Weller DM. Biological control of soilborne plant pathogens with rhizosphere bacteria. Ann. Rev. Phytopathol. 1988;26:379-407.
Crossref - Watanabe T, Suzuki K, Oyanagi W, Ohnishi K, Tanaka H. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J Biolog Chem. 1990;265(26):15659-15665.
Crossref - Chang WT, Chen ML, Wang SL. An antifungal chitinase produced by Bacillus subtilis using chitin waste as a carbon source. World J Microbiol Biotechnol. 2010;26(5):945-950.
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN (ed.), Mammalian Protein Metabolism. Academic Press, New York. 1969;21-132.
Crossref - Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
- Imoto T, Yagishita K. A simple activity measurement by lysozyme. Agric Biol Chem. 1971;35(7):1154-1156.
Crossref - Sambrook J, Fritsch EF, Maniatis T. Cold spring harbor laboratory press; New York: Molecular cloning: a laboratory manual. 1989:9-14.
- Abdelsalam IS. Production of Vitamin B12 and Folate Using a Potent Mutant Strain of Klebsiella pneumonia. Egypt J Chem. 2018;61(1):93-100.
Crossref - Khusro A, JPR Raj, Panicker SG. Study on antagonistic activity of a novel bacterial isolate under mild stress condition of certain antimicrobial agents. Eur J Exp Biol. 2014;4(4):26-30.
- Aunpad R, Na-Bangchang K. P2085 A novel antimicrobial peptide with anti-MRSA activity. Int J Antimicrob Agents. 2007;(29):S602.
- Yadav V, Mandhan R, Kumar M, Gupta J, Sharma GL. Characterization of the Escherichia coli antifungal protein PPEBL21. Int J Microbiol 2010;2010:196363.
Crossref - Yadav V, Mandhan R, Pasha Q, et al. An antifungal protein from Escherichia coli. J Med Microbiol. 2007;56(5):637-644.
Crossref - Ramachandran R, Chalasani AG, Lal R, Roy U. A broad-spectrum antimicrobial activity of Bacillus subtilis RLID 12.1. The Scientific World Journal. 2014;2014:1968487.
Crossref - Li X, Zhang Y, Wei Z, Guan Z, Cai Y, Liao X. Antifungal activity of isolated Bacillus amyloliquefaciens SYBC H47 for the biocontrol of peach gummosis. PloS One. 2016;11(9):e0162125.
Crossref - Mardanova AM, Hadieva GF, Lutfullin MT, et al. Bacillus subtilis strains with antifungal activity against the phytopathogenic fungi. Agricultural Sciences. 2017;8(1):1-20.
Crossref - Mushtaq S, Ali A, Khokhar I, Mukhtar I. Antagonisitic potential of soil bacteria against food borne fungi. World Appl Sci J. 2010;11(8):966-969.
- Senol M, Nadaroglu H, Dikbas N, Kotan R. Purification of Chitinase enzymes from Bacillus subtilis bacteria TV-125, investigation of kinetic properties and antifungal activity against Fusarium culmorum. Ann Clin Microbiol Antimicrob. 2014;13(1):35.
Crossref - Sreeju SN, Babu MM, Mariappan C, Selvamohan T. Effect of physical and chemical mutagens on biopolmer producing strains and RAPD analysis of mutated strains. Arch Appl Sci Res. 2011;3(6):233-246.
- Patel RM, Sudha. Mutation detection in Bacillus amyloliquefaciens by random amplified polymorphic DNA-(RAPD) PCR. Pharma Research. 2013;9(1):13-21.
- Khattab AA, Ghafar HHA, Ibrahim MIM. Enhancement of phenol biosorption by genetically improved of Rhodosporidium toruloides strains. J Appl Sci Res. 2012:3600-3607.
- Gomaa EZ, El-Mahdy OM. Improvement of Chitinase Production by Bacillus thuringiensis NM101-19 for Antifungal Biocontrol through Physical Mutation. Microbiology. 2018;87(4):472-485.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.