ISSN: 0973-7510
E-ISSN: 2581-690X
Antimicrobial therapy is frequently associated with the emergence of resistant bacteria with a high rate of morbidity and mortality worldwide. The present study was aimed at investigating the impact of a neodymium-doped yttrium aluminum (Nd:YAG) laser, and a static magnetic field (SMF) on cellular growth and DNA alteration in some clinical bacterial isolates. Samples from cutaneous wounds were collected by sterile cotton swabs from three elderly women admitted to Tikrit Teaching Hospital, Tikrit City, Iraq. Isolation and identification of Streptococcus agalactiae, Staphylococcus aureus, and Pseudomonas aeruginosa were carried out using cultural characteristics, microscopy, and biochemical tests. Three broth cultures were prepared for each of the test isolates. The first broth culture served as untreated control, the second was exposed to an Nd:YAG laser and the third was exposed to SMF. Colony counting was done on all the samples. DNA was extracted from the test bacteria and used to perform the RAPD-PCR assay. In contrast to the untreated control, the results showed that Nd:YAG laser radiation was more effective than SMF at inhibiting the cellular growth of the test isolates. Also, the radiation caused DNA alteration, which was established by decreased microbial growth, as well as the development of new bands and the loss of original bands. According to the findings of this study, the Nd:YAG laser is a promising technique for influencing the healing of infected cutaneous wounds. RAPD-PCR is also a useful biomarker assay for assessing the biological impact of laser radiation and SMF on bacteria.
Nd:YAG Laser, Static Magnetic Field, RAPD-PCR, S. agalactiae, S. aureus, Pseudomonas aeruginosa
Antimicrobial therapy is frequently associated with the emergence of resistant bacteria, which is one of the largest problems and risks to global health.1-3 Consequently, it is urgently needed to accelerate research and development for new antimicrobial strategies,4 as well as for quick infection control during wound healing.5 Laser radiation, a physical therapy modality with some advantages, including painlessness, non-invasiveness, and the ability to treat a variety of pathological diseases, has recently been used to control microbial activity.6 Additionally, the healing of soft tissue wounds that could harbor bacterial colonization was demonstrated by the therapeutic action of low-intensity laser irradiation.7 Some researchers have shown that pulsed high-intensity neodymium-doped yttrium aluminum garnet (Nd:YAG [Nd:Y3Al5O12]) laser has antibacterial properties.8,9 The effects of laser irradiation on Candida albicans, Staphylococcus aureus, and Pseudomonas aeruginosa have been established.10-12 Nd:YAG laser is a high-intensity laser therapy that transmits light in the infrared area at a wavelength of 1,064 nm, which permits it to diffuse into the tissue.6 Water molecules within microbes absorb laser photons, causing genetic modification and killing pathogenic microorganisms. This also slows or stops microbial growth.13 However, the effect of the laser on living cells is related to the time of exposure, energy, and wavelength of the laser beam.14 Static magnetic field (SMF) represent another strategy to control microbial growth concerning its biological effect on different microbial cells in term of inhibiting cellular growth,15-17 as well as genetic expression.18,19 According to the recommendation of the European Union, which Italy enacted into law, SMF below 0.5 T are completely safe for humans, and no license is required for using devices with SMF less than 0.5 T, such as in MR tomography.19
Recently, several sensitive molecular assays have been developed for the detection of DNA mutation or damage by comparing DNA profiles from untreated control with genotoxic agent-treated samples.20 Some of these assays include repetitive sequence-based polymerase chain reaction (PCR),18 RNA arbitrarily primed PCR,19 and random amplified polymorphic DNA- (RAPD)-PCR assays.9 The RAPD-PCR method is used to amplify bands of template DNA using arbitrary primers, typically of 10 nucleotides in length. The primer anneals to genomic DNA with a nearly identical complementary sequence. If the two priming sites are close to one another, amplification occurs.21 RAPD-PCR is a technique for studying DNA patterns in various species by detecting changes in the intensity of amplified bands, the emergence of new bands, and the loss of preexisting bands after toxicant exposure.22 This technique is reliable and very sensitive and does not require radioisotopes, or previous knowledge of the genome sequence of the test organism.23 It can show changes in the genomic DNA as a whole instead of the precise location.24 Consequently, this technique attracts the attention of many researchers and is used in a variety of biological fields, such as epidemiological studies to type different organisms,25-27 genomic instability of cancer,28 detections of genomic mutation brought on by genotoxic material, such as chemical agents,29 gamma radiation, 30 and laser radiation.9
The present study was conducted to assess the impact of the Nd:YAG laser and SMF on cellular growth and DNA alteration of some bacterial species using colony counting technique and the RAPD-PCR assay, respectively.
Sample Collection
Clinical samples from cutaneous wounds were collected by sterile cotton swabs from three elderly women who were brought to the Tikrit Teaching Hospital in Tikrit City, Iraq in January 2021. The samples were immediately sent to the microbiological laboratory, Department of Biology, College of Science, University of Tikrit, Tikrit, Iraq.
Microbiological Analysis of the Clinical Samples
The samples were cultured on different media include: nutrient agar, blood agar, and mannitol agar (Hemedia, India). In addition to Kirby-Bauer agar for detection susceptibility of S. agalactiae to bacitracin (Hemedia, India)Consequently, the isolated bacteria were identified based on colony morphology, measurement of hemolytic activity on 5% sheep blood agar, gram staining, oxidase, catalase, motility, and conventional biochemical tests for the identification of S. agalactiae, S. aureus, and Pseudomonas aeruginosa.31 Then, the identification was confirmed by using the VITEK 2 Compact System (BioMeriex, Inc., France) according to the manufacturer’s instructions.
Experimental Groupings and Treatment
From each isolate on the selected media, one colony of bacteria was picked up, added to the brain heart infusion broth (Hemedia, India), and centrifuged for 10 minutes at 6000 rpm. The pellets were combined using a vortex and suspended in 5ml of normal saline after being rinsed several times with the saline solution. Each suspended isolate was then serially diluted and then incubated at 37°C for 24 hours. The total viable count was performed on the plates containing 30 – 300 bacterial colonies. Nine broth cultures containing 1 ml of suitable dilution of each bacterial suspension were prepared and then divided into three groups. The first group served as the untreated control; the second group was exposed to an Nd:YAG laser using a 1,064 nm wavelength, 500 mJ energy, 5Hz frequency, 20 second exposure time, and a 10 cm distance from the laser device (Germany). The third group was exposed to SMF produced by a locally constructed dipolar magnet (500 mT, 20-second exposure time), with its force being measured by a teslameter at the Department of Physical Science, College of Science, University of Tikrit. Following a 24-hour incubation period and exposure to the Nd:YAG laser and SMF, colony counting procedures were conducted on all treated and untreated samples. Later, the mean value for each group was computed.32
DNA Extraction from Clinical Samples
Streptococcus aureus and S. agalactiae were grown on tryptic soy agar supplemented with 5% human blood, while P. aeruginosa isolates were cultured on nutrient agar. The cultures were incubated at 37˚C for 24 hours. Streptococcus agalactiae and S. aureus DNAs were extracted using the Wizard genomic DNA extraction kit (Promega, USA) following the manufacturer’s instructions and stored at -20°C. Meanwhile, DNA was extracted from P. aeruginosa according to the method described by Chen and Kuo.33 The isolated DNA from each species was diluted, and its purity and concentration were determined by measuring absorbance at 260 and 280 nm using Nanodrop (Thermo Scientific, Germany) before the PCR amplification procedure was performed.
RAPD-PCR Assay
DNA was amplified using two 10-mer primers (Operan Technologies, Inc. in the USA): OPH-08, with a sequence of 5′-GAAACACCCC-3′, and OPI-02, with a sequence of 5′-(GGAGGAGAGG-3′.34 The RAPD-PCR was prepared using the Accu Power PCR premix (Pioneer, Korea) following the manufacturer’s protocol. The DNA template (50 ng) and 10 pmole of primer were added to the master mix, which also contained 250 µm of dNTPS, 1U of Taq DNA polymerase, 10 mM of Tris-HCl (pH 9), 30 mM of KCl, and MgCl2. Then, the final volume was made up to 20 µl by adding distilled water. The amplification program started with one cycle of initial denaturation at 94˚C for 2 minutes, followed by 40 cycles of denaturation at 92˚C for 1 minute, annealing at 37˚C for 1 minute, elongation at 72˚C for 1 minute, and final elongation at 72˚C for 7 minutes. The amplified DNA of test isolates was separated using 1.5% agarose gel electrophoresis in 1xTBE for 1 hour (5 Volts/cm), together with a standard molecular marker, then stained with ethidium bromide and visualized under a UV trans illuminator.35 The colony count for the control and treated samples, as well as the percentage of colony count reduction after 24 hours of exposure to Nd:YAG laser and SMF, were calculated using the following equation:
Percentage colony count = [ Colony count (control) – colony count after 24 hrs (experimental) / Colony count ] × 100
Percentage polymorphism bands were calculated using the following equation:36
Polymorphism bands (%) = [Total No. of a + b bands / Total No. of control bands] × 100
Where
a: number of new appeared bands;
b: number of disappeared bands
Statistical Analysis
Three replicates were used for each type of treated bacteria then, the mean and standard deviation of number of colonies for each treated sample were calculated using the Statistical Package for the Social Sciences (SPSS) version 22.0 software.
Streptococcus agalactiae, S. aureus, and P. aeruginosa were the three bacterial species that were recovered from clinical samples based on morphological characteristics and cultural properties as well as conventional biochemical tests. The non-motile, gram-positive cocci that were grouped in clusters, gave a negative reaction to the oxidase test, were coagulase-positive, fermented mannitol, and produced β-hemolysis on blood agar were identified as S. aureus. On the other hand, S. agalactiae was identified as the gram-positive oval or spherical-shaped cocci that were arranged in pairs or chains and produced a negative reaction to the oxidase, catalase, and coagulase tests, and bile esculin agar, did not ferment mannitol, were not motile, resistance to bacitracin and produce a hemolytic reaction on 5% sheep blood agar. Meanwhile, P. aeruginosa was identified as gram-negative motile rods, that produce oxidase, catalase, and β- hemolysis on blood agar, giving a positive citrate utilization reaction, but negative for indole, methyl red, and Voges–Proskauer tests. They grew at 42°C, with a colony that was blue-green colored and had a grape-like odor on cetrimide agar. Later, these identifications were confirmed with VITEK 2 compact system (BioMeriex, Inc., France).
Table (1):
The mean ± standard deviation values of colony counting for controls and experimental samples of test bacterial isolates after exposure to Nd:YAG laser and SMF.
Bacteria |
M ±SD of C |
M±SD of Laser treated samples |
% ↓ No. of colony after Laser treatment |
M±SD of No. of colony SMF after treatment |
% ↓ in No. of colony after SMF treatment |
|---|---|---|---|---|---|
S. agalactiae |
195±05 |
142±05 |
27.18 |
165±07 |
15.38 |
S. aureus |
181±11 |
153±09 |
15.47 |
167±33 |
7.73 |
P. aeruginosa |
292±04 |
270±04 |
7.53 |
278±02 |
4.79 |
↓ : decrease, C: Control; M: Mean; Nd:YAG: Neodymium-doped yttrium aluminum laser; SMF: Static magnetic field, %: percentage
Table 1 demonstrated the mean value and standard deviation of colony count of the control and treated samples of the test bacterial isolates after exposure to Nd:YAG laser and SMF for S. agalactiae (195,142, and 165), S. aureus (181,153, and 167), and P. aeruginosa (292,270, and 278). For the three bacterial species, S. agalactiae, S. aureus, and P. aeruginosa, the percentages of colony reduction following laser therapy (500 mJ, 20 s) were 27.18, 15.47, and 7.53, respectively. Meanwhile, the test isolates’ percentages of colony reduction following SMF treatment (500 mT, 20 seconds) were 15.38, 7.73, and 4.79, respectively. These results showed that SMF has antibacterial properties, and the Nd:YAG laser has bactericidal properties. These effects were found to be statistically significant. However, it was observed that the effectiveness of Nd:YAG laser was stronger than the SMF influence on the test isolates, and they have less impact on the growth of P. aeruginosa.
Table (2):
RAPD-PCR profiles produced from genomic DNA of test bacteria after treatment with Nd:YAG laser and SMF.
| Primer code | Bacterial species | Nd. YAG Laser treated samples | MF treated samples | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | T | P | M | a | b | T | P | M | a | b | ||
| OPI-02 | S. agalactiae | 5 | 7 | 6 | 3 | 4 | 2 | 5 | 6 | 2 | 3 | 3 |
| S. aureus | 3 | 2 | 3 | 1 | 1 | 2 | 4 | 5 | 1 | 3 | 2 | |
| P. aeruginosa | 6 | 4 | 6 | 1 | 2 | 4 | 6 | 2 | 5 | 1 | 1 | |
| Total | 14 | 12 | 13 | 5 | 7 | 8 | 15 | 13 | 8 | 7 | 6 | |
| a+b | 15 | 13 | ||||||||||
| Polymorphic (%) | 107.14 | 100 | ||||||||||
| Total Polymorphic (%) | 207.14 | |||||||||||
| OPH-8 | Bacterial species | C | T | P | M | a | b | T | P | M | a | b |
| S. agalactiae | 5 | 5 | 10 | 0 | 5 | 5 | 5 | 8 | 1 | 4 | 4 | |
| S. aureus | 4 | 2 | 6 | 0 | 2 | 4 | 2 | 6 | 0 | 2 | 4 | |
| P. aeruginosa | 2 | 4 | 6 | 0 | 4 | 2 | 2 | 2 | 1 | 1 | 1 | |
| Total | 11 | 11 | 22 | 0 | 11 | 11 | 9 | 16 | 2 | 7 | 9 | |
| a+b | 22 | 16 | ||||||||||
| Polymorphic (%) | 200 | 177.8 | ||||||||||
| Total Polymorphic (%) | 377.8 | |||||||||||
RAPD-PCR: Random amplified polymorphic DNA-polymerase chain reaction; C: Control; T: Total number of Nd:YAG laser treated bands; P: Polymorphic bands; M: Monomorphic bands; a: New appearing band; b: Disappearing band; T SMF: Total number of SMF treated bands.
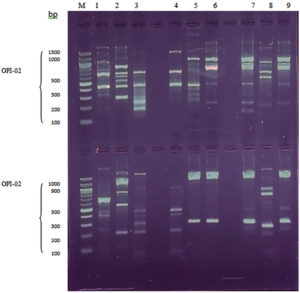
As depicted in Table 2 and Figure, both the RAPD-PCR primers yielded amplified products. These amplified products were highly polymorphic and reproducible and produced a total of 73 bands with 66 polymorphic bands across all tested isolates, including 25 bands of controls. The molecular size of the RAPD bands ranged approximately between 150-2300 bp. Concerning the OPH-08 RAPD-PCR primer, it produced a total of 42 bands of DNA extracted from the three test bacterial isolates, with molecular sizes ranging from 150 to 2,300 bp. There were (13, 15), (14), and (14), bands, respectively, for the laser-treated, SMF-treated, and control bacterial isolates, as well as (15), and (13), polymorphic bands for Nd:YAG laser and SMF treated samples, respectively. Furthermore, seven new bands appeared for each laser and SMF-treated sample. Meanwhile, for samples treated with the Nd:YAG laser and SMF, (8 and 6) bands, respectively, disappeared. The OPI-02 primer amplified a total of 31 bands across all the test DNA samples, including (11, 9, and 11) bands of laser-treated, SMF treated, and control samples, respectively, including (22 and 16) polymorphic bands of Nd:YAG laser and SMF treated samples, respectively. There were (11 and 7) new bands that appeared with the disappearance of (11, and 9) bands for samples exposed to Nd:YAG laser and SMF, respectively. These results revealed a considerable alteration in the number and molecular size. However, the RAPD-PCR profile produced by primer OPI-02 generated more polymorphic bands than that of OPH-08.
Figure. RAPD–PCR profile of genomic DNA from bacteria exposed to Nd: YAG laser and SMF. Lane (M): Molecular marker; Lane 1: Untreated control S. agalactiae; Lane 2: Laser treated S. agalactiae; Lane 3: SMF treated; Lane 4: Untreated control S. aureu; Lane 5: Laser treated S. aureus; Lane 6: SMF-treated S. aureus; Lane 7 untreated control P. aeruginosa; Lane 8: Laser-treated P. aeruginosa; Lane 9: SMF-treated P. aeruginosa
The comparative effect of the Nd:YAG laser and SMF treatment on the three test isolates (S. agalactiae, S. aureus, and P. aeruginosa) is shown in Table 3. A total of (16, 9, and 12) polymorphic bands were generated by the RAPD-PCR assay using the OPH-08 and OPI-02 primers on DNA samples extracted from S. agalactiae, S. aureus, and P. aeruginosa, respectively. In contrast, DNA samples taken after SMF exposure from S. agalactiae, S. aureus, and P. aeruginosa, respectively, produced (14), (12), and (4) polymorphism bands. This result made it very evident that Nd:YAG laser, as opposed to SMF force, had a greater impact on S. agalactiae and P. aeruginosa. In contrast, there were (14, 12, and 4) polymorphic bands generated from DNA samples extracted from S. agalactiae, S. aureus, and P. aeruginosa, respectively, after SMF exposure. This result indicated that Nd:YAG laser had a greater impact on S. agalactiae and P. aeruginosa than SMF force. Additionally, the two primers produced more bands when they amplified DNA samples from S. agalactiae after subjecting them to an Nd:YAC laser and SMF force treatment. However, the DNA amplified product employing OPI-02 primer on DNA extracted from P. aeruginosa that was exposed to SMF did not exhibit any differences from the control. This result demonstrated that SMF had less of an impact on this bacterial species.
Table (3):
RAPD PCR profile of genomic DNA from test bacteria after treatment with Nd:YAG laser and SMF.
| Bacteria | Variable | Primer code | C | Nd. YAG Laser treated samples | MF samples | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | P | M | a | b | T | P | M | a | b | ||||
| S. agalactiae | OPI-02 | 5 | 7 | 6 | 3 | 4 | 2 | 5 | 6 | 2 | 3 | 3 | |
| OPH-8 | 5 | 5 | 10 | 0 | 5 | 5 | 5 | 8 | 1 | 4 | 4 | ||
| Total | 10 | 12 | 16 | 3 | 9 | 7 | 10 | 14 | 2 | 7 | 7 | ||
| a+b | 16 | 14 | |||||||||||
| P (%) | 160 | 140 | |||||||||||
| Total P (%) | 300 | ||||||||||||
| S. aureus | OPI-02 | 3 | 2 | 3 | 1 | 1 | 2 | 4 | 5 | 1 | 3 | 2 | |
| OPH-8 | 4 | 2 | 6 | 0 | 2 | 4 | 2 | 6 | 0 | 2 | 4 | ||
| T | 7 | 4 | 9 | 1 | 3 | 6 | 6 | 11 | 1 | 5 | 6 | ||
| a+b | 9 | 11 | |||||||||||
| P (%) | 128.57 | 157.14 | |||||||||||
| Total P (%) | 285-71 | ||||||||||||
| P. aeruginosa | OPI-02 | 6 | 4 | 6 | 1 | 2 | 4 | 6 | 2 | 5 | 1 | 1 | |
| OPH-8 | 2 | 4 | 6 | 0 | 4 | 2 | 2 | 0 | 2 | 0 | 0 | ||
| T | 8 | 8 | 12 | 1 | 6 | 6 | 8 | 2 | 7 | 1 | 1 | ||
| a+b | 12 | 2 | |||||||||||
| P (%) | 150 | 25 | |||||||||||
| Total P (%) | 175 | ||||||||||||
C: Control; T: Total number of Nd:YAG laser treated bands; P: Polymorphic bands; M: Monomorphic bands; a: New appearing band; b: Disappearing band.
The percentage of colony count was reduced after Nd:YAG laser and SMF treatments for all the test isolates, S. agalactiae, S. aureus, and P. aeruginosa, as compared to untreated control samples, demonstrating that Nd:YAG laser and SMF have an inhibitory effect on cell growth (Table 1). However, the bactericidal effect of the Nd:YAG laser was greater than that of SMF. This observation agrees with what other researchers had previously reported.8 They discovered a significant decrease in colony count in all test samples of P. aeruginosa and S. aureus following 24 hours of pulsed high-intensity Nd:YAG laser irradiation for all doses (500, 600, 700, and 800 J). Furthermore, as the irradiation doses were raised, the effect directly increased. However, Bergmans et al. demonstrated that Nd:YAG laser irradiation of inoculated Enterococcus faecalis into root canals generated a significant reduction in E. faecalis cell counts, resulting in 97.3% fewer bacteria, as well as disrupting biofilm and cell structure. They concluded that Nd:YAG laser irradiation is a complement and not an alternative to various disinfection regimens in chronic infections of pathogenic bacteria like E. faecalis that are resistant to several antimicrobial treatments.37 In another study, after receiving a second session with a long-pulsed Nd:YAG laser, 78.6% of the 145 patients who had vessels on their faces or legs showed excellent clinical improvement. The most common side effect observed in 12.4% of all participants was erythema.38
Kim et al.,39 also investigated the efficacy and safety of removing melanocytic nevi with a 1,064 nm Q-switched Nd: YAG laser. A total of 2,064 Korean patients participated in this study. They reported its effectiveness, and safety in the treatment of all the participants, except for minor side effects like reepithelization, redness, and prickling during treatment, no infection after surgery, moderate post-treatment erythema, and swelling. All of these adverse effects disappeared within a few hours to two weeks. Also, treatment-induced pinpoint bleeding lasted three to five days after treatment. Investigations were also done into how different bacterial species were affected by magnetic fields. Masood,40 reported that a weak static magnetic field of a few gausses for a few hours causes a significant change in the growth rates of Escherichia coli, P. aeruginosa, and S. epidermidis, and demonstrated that the magnetic field has diverse effects on these different species in the same environmental setting. After 24 hours of exposure, a magnetic field also slows the growth of the bacteria E. coli and S. aureus, while speeding up the growth of Bacillus subtilis.17 In addition, neither S. epidermidis nor S. aureus showed a statistically significant reduction in cell proliferation when exposed to electromagnetic fields, except for S. aureus when exposed to 900 MHz for 12 hours, which was cultured in media containing 30g of amoxicillin before being exposed to electromagnetic fields. While P. aeruginosa cell growth rate was unaffected by 900 MHz electromagnetic fields.18 All biological systems depend on differential gene expression, which enables both the adaptation to challenging environmental conditions and the proper and appropriate responses under normal conditions. Microbiological cells may experience stress or toxicity as a result of irradiation with Nd:YAG laser and SMF, which may affect their ability to grow and express their genetic material.
The RAPD-PCR technique represents a molecular tool used to identify and isolate genes that are differentially expressed in living cells.41 In this study, three bacterial species; S. agalactiae, S. aureus, and P. aeruginosa isolated from cutaneous wound samples were selected for determining any possible mutation or alteration in genomic DNA following exposure to Nd:YAG laser and SMF by applying a RAPD-PCR assay using two random primers (OPH-08, and OPI-02). The reproducibility of this technique was assessed by repeating the procedure three times for each primer tested. Analysis of the results was based on counting the total number of bands amplified, the “total number of polymorphic bands,” which represents the number of new bands that are introduced, and the number of bands that are absent after exposure to the Nd:YAG laser and SMF in comparison to the results of the untreated control. The result of this study demonstrated that the two kinds of primers utilized (OPH-08 and OPI-02) produced different banding patterns in the isolated bacterial DNA. Depending on the primers used, the quality of the genetic pattern, or the availability of different primer binding sites in the genome, the number of bands may change. Some primers seem to function more effectively than others and can produce consistent results.42 In comparison to an unexposed control, the RAPD approach was shown to be effective in differentiating the test isolates and assessing genetic diversity after exposure to Nd:YAG laser and SMF. This observation revealed the feasibility and applicability of RAPD-PCR in such an investigation, which mostly depends on the selection of primers that result in amplification patterns. In this regard, multiple studies have demonstrated the benefit of the RAPD-PCR method in comparing changes in genetic expression following exposure to various carcinogenic agents or radiation.29,43 Some researchers successfully used the RAPD-PCR method to improve the impact of the Nd:YAG laser with a wavelength of 1,064 nm (300 and 500 mJ energies for 15 and 25 seconds) on the genetic material of P. aeruginosa.9 Their report was found to agree with the findings of the current study, despite differences in the energy and duration of application. Other researchers discovered that the Nd:YAG laser and diode had an impact on the genomic DNA of Candida albicans and changed the RAPD-PCR profile significantly when compared to the control group.44 Laser therapy can inhibit the physiological activity of microbial cells by suppressing DNA metabolism, degenerative changes, alteration in cell division, cytomorphology, and cell cytokinesis. The level of cellular destruction varies depending on laser type, laser parameters, and doses.45 Ishikawa et.al.46 reported that Nd:YAG laser radiation has a promising future when used as an adjunct to conventional mechanical nonsurgical therapy.
SMF caused changes in the RAPD-PCR profile of the amplified DNA samples of the test isolates. However, it had a negligible impact on P. aeruginosa. According to Potenza et al., SMF exposure caused changes in gene expression in E. coli and may have stimulated transposition activity.47 They also hypothesized that magnetic fields have genotoxic effects on living organisms and can disrupt DNA stability either directly through interactions with DNA or through the activity of oxidizing radicals. Magnet characteristics, magnet support device, target tissue(s), and dosing regimen are SMF parameters that are important for interaction with a biological system.48 However, the biological effects of SMF are significantly influenced by the field gradient and intensity.49,50 It was determined that regardless of the magnetic density, exposure to SMFs alone has no or very minor effects on cell development and genetic damage.51 According to the findings of Colbert et al., before beginning a large-scale clinical trial, it is essential to optimize SMF dose parameters for specific clinical conditions.52
The findings of this study reveal that the Nd:YAG laser is a promising technology for influencing the healing of infected cutaneous wounds, and that its effects outweigh those of SMF on antibiotic-resistant isolates of the bacteria, S. agalactiae, S. aureus, and P. aeruginosa. The RAPD-PCR biomarker assay is also useful for assessing the biological effects of radiation and static magnetic fields on bacteria. It is recommended that the antibacterial activity of SMF and Nd:YAG laser on other pathogenic bacteria be investigated.
ACKNOWLEDGMENTS
The authors would like to thank their respective universities for providing the necessary support to conduct this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Ethics Committee of the College of Science, University of Tikrit, and College of Nursing, University of Kirkuk, Iraq with reference number (30/7/1874) on 18/11/2021.
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st centurya clinical super-challenge. N Engl J Med. 2009;360(5):439-443.
Crossref - Karim GF, AL-Salihi SS, Atya QM, Abass KS. Aerobic and Anaerobic Bacteria in Tonsils of Different Ages with Recurrent Tonsillitis. Indian J Public Health Res Dev. 2019;10(9):132-136.
Crossref - Raabe VN, Shane AL. Group B Streptococcus (Streptococcus agalactiae). Microbiol Spectr. 2019;7(2):10.1128.
Crossref - Bai H, He W, Chau JHC, et al. AIEgens for microbial detection and antimicrobial therapy. Biomaterials. 2021;268:120598.
Crossref - Athanasiou A, Karkambounas S, Batistatou A, et al. The effect of pulsed electromagnetic fields on secondary skin wound healing: an experimental study. Bioelectromagnetics. 2007;28(5):362-368.
Crossref - Ebid AA, Ibrahim AR, Omar MT, El Baky AMA. Long-term effects of pulsed high-intensity laser therapy in the treatment of post-burn pruritus: a double-blind, placebo-controlled, randomized study. J Lasers Med Sci. 2017;32(3):693-701.
Crossref - de Sousa NTA, Gomes RC, Santos MF, Brandino HE, Martinez R, de Jesus Guirro RR. Red and infrared laser therapy inhibits in vitro growth of major bacterial species that commonly colonize skin ulcers. J Laser Med Sci. 2016;31(3):549-556.
Crossref - Ebid AA, Alhammad RM, Alhendi RT, et al. Immediate effect of pulsed high-intensity neodymium-doped yttrium aluminum garnet (Nd: YAG) laser on staphylococcus aureus and pseudomonas aeruginosa growth: an experimental study. J Phys Ther Sci. 2019;31(11):925-930.
Crossref - Al-Doori AA, Jasem AS, Al-Azzawie AF. Assessment of the molecular effects of Nd: YAG laser on Pseudomonas aeruginosa bacteria using random amplified polymorphic DNA marker. Drug Invention Today. 2020;14(2):308-331. https://www.researchgate.net/publication/340398078_Assessment_of_
the_molecular_effects_of_Nd_YAG_laser_on_Pseudomonas_aeruginosa_bacteria
_using_random_amplified_polymorphic_DNA_marker - Grzech-Lesniak K, Nowicka J, Pajaczkowska M, et al., Effects of Nd: YAG laser irradiation on the growth of Candida albicans and Streptococcus mutans: in vitro study. J Laser Med Sci. 2019;34(1):129-137.
Crossref - Al-Taie KLS, Rasheed HM. Effects of Nd: YAG (1064NM) on staphylococcus aureus isolated from wounds. Plant Archives. 2019;19(2):1728-1731. http://plantarchives.org/SPL%20ISSUE%20SUPP%202,2019/300__1728-1731_.pdf
- Ebid AA, Alhammad RM, Alhindi RT, et al. Effect of high-power Nd: YAG laser on the growth of Staphylococcus aureus and Pseudomonas aeruginosa: an experimental study. J Phys Ther Sci. 2021;33(3):222-228.
Crossref - Machado RS, Viana S, Sbruzzi G. Low-level laser therapy in the treatment of pressure ulcers: systematic review. Lasers Med Sci. 2017;32(4):937-944.
Crossref - Hawkins D, Houreld N, Abrahamse H. Low level laser therapy (LLLT) as an effective therapeutic modality for delayed wound healing. Ann N Y Acad. Sci. 2005;1056(1):486-493.
Crossref - Haagensen JA, Bache M, Giuliani L, Blom NS. Effects of Resonant Electromagnetic Fields on Biofilm Formation in Pseudomonas aeruginosa. Appl Sci. 2021;11(16):7760.
Crossref - Mousavian-Roshanzamir S, Makhdoumi-Kakhki A. The inhibitory effects of static magnetic field on Escherichia coli from two different sources at short exposure time. Rep Biochem Mol Biol. 2017;5(2):112-116.PMID: 28367473
- Al-Khaza’leh KA, Al-fawwaz AT. The Effect of Static Magnetic Field on E. coli, S. aureus and B. subtilis Viability. J Nat Sci Res. 2015;5(2):153-157. PMCID: PMC5346279
- Salmen SH, Alharbi SA, Faden AA, Wainwright M. Evaluation of effect of high frequency electromagnetic field on growth and antibiotic sensitivity of bacteria. Saudi J Biol Sci. 2018;25(1):105-110.
Crossref - Potenza L, Ubaldi L, De Sanctis R, De Bellis R, Cucchiarini L, Dacha M. Effects of a static magnetic field on cell growth and gene expression in Escherichia coli. Mutat Res. 2004;561(1-2):53-62.
Crossref - Enan MR. Application of random amplified polymorphic DNA (RAPD) to detect the genotoxic effect of heavy metals. Biotechnol Appl Biochem. 2006;43(3):147-154.
Crossref - Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;25;18(24):7213-7218.
Crossref - Atienzar FA, Venier P, Jha AN, Depledge MH. Evaluation of the random amplified polymorphic DNA (RAPD) assay for the detection of DNA damage, and mutations. Mutat Res. 2002;521(1-2):151-163.
Crossref - Atienzar FA, Jha AN. The random amplified polymorphic DNA (RAPD) assay and related techniques applied to genotoxicity and carcinogenesis studies: a critical review. Mutat Res. 2006;613(2-3):76-102.
Crossref - Sorkh MAG, Shokoohizadeh L, Rashidi N, Tajbakhsh E. Molecular analysis of Pseudomonas aeruginosa strains isolated from burn patients by repetitive extragenic palindromic-PCR (rep-PCR). ARCMJ. 2017;19(4):e43508.
Crossref - Parsa P, Amirmozafari N, Nowruzi B, Bahar MA. Molecular characterization of polymorphisms among Pseudomonas aeruginosa strains isolated from burn patients’ wounds. Heliyon. 2020;6(12):e05041.
Crossref - Amal MNA, Zamri-Saad M, Siti-Zahrah A, Zulkafli AR, Nur-Nazifah M. Molecular characterization of Streptococcus agalactiae strains isolated from fishes in Malaysia. J of Appl Microbiol. 2013;115(1):20-29.
Crossref - Al-Assie AH, Al-Azawy AF, Al-Dori WS. Molecular characterization of Pseudomonas aeruginosa Isolates Isolated from Clinical Patients by Using RAPD-PCR Technique. J of University of Anbar for Pure Science. 2012;6(3). https://www.iasj.net/iasj/pdf/09598797483f3f48
- Papadopoulos S, Benter T, Anastassiou G, Pape M, Gerhard S, Bornfeld N, Dorken B. Assessment of genomic instability in breast cancer and uveal melanoma by random amplified polymorphic DNA analysis. Int J Cancer. 2002;99(2):193-200.
Crossref - Dogan I, Ozyigit II, Tombuloglu G, Sakcali MS, Tombuloglu H. Assessment of Cd-induced genotoxic damage in Urtica pilulifera L. using RAPD-PCR analysis. Biotechnol Biotechnol Equip. 2016;30(2):284-291.
Crossref - Hegazi AZ, Hamideldin N. The effect of gamma irradiation on enhancement of growth and seed yield of okra [Abelmoschus esculentus (L.) Monech] and associated molecular changes. Journal of Horticulture and Forestry. 2010;2(3):38-51.
https://www.researchgate.net/publication/259461159_The_effect_of_gamma_
irradiation_on_enhancement_of_growth_and_seed_yield_of_okra_Abelmoschus
_esculentus_L_Monech_and_associated_molecular_changes - Fischbach FT, Dunning MB. A manual of laboratory and diagnostic tests. Philadelphia. 2009. Wolters Kluwer Health/Lippincott Williams & Wilkins.
- Abo-Ksour, Mohammed F.; Al-Jubori, Sawsan S.; Jawad, Hussein A. Influence of Helium-Neon Laser on Some Virulence Factors of Staphylococcus Aureus and Escherichia Coli. Al-Mustansiriyah Journal of Science, 2018, 29.3: 29-34.
- Chen WP, Kuo TT. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 1993;21(9):2260.
Crossref - Onasanya A, Mignouna HD, Thottappilly G. Genetic fingerprinting and phylogenetic diversity of Staphylococcus aureus isolates from Nigeria. Afr J Biotechnol. 2003;2(8):246-250.
Crossref - Weigand F, Baum M, Udupa S. DNA molecular marker techniques. Technical Manual No. 20″@eng. ICARDA. 1993.
- Taspinar MS, Agar G, Yildirim N, Sunar S, Aksakal O, Bozari S. Evaluation of selenium effect on cadmium genotoxicity in Vicia faba using RAPD. J Food Agric Environ. 2009;7 (3&4):857-860.
- Bergmans L, Moisiadis P, Teughels W, Van Meerbeek B, Quirynen M, Lambrechts P. Bactericidal effect of Nd: YAG laser irradiation on some endodontic pathogens ex vivo. Int Endod J. 2006;39(7):547-557.
Crossref - Serdar ZS, Izci NF. The evaluation of long-pulsed Nd:YAG laser efficacy and side effects in the treatment of cutaneous vessels on the face and legs. J Cosmet Dermatol. 2020;19(7):1656-1661.
Crossref - Kim YJ, Whang KU, Choi WB, et al. Efficacy and safety of 1,064 nm Q-switched Nd:YAG laser treatment for removing melanocytic nevi. Ann Dermatol. 2012;24(2):162-167.
Crossref - Masood S. Effect of weak magnetic field on bacterial growth. Biophys Rev. 2017;12(04):177-186.
Crossref - Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257(5072):967-971.
Crossref - Tyler KD, Wang G, Tyler SD, Johnson WM. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol. 1997;35(2):339-346.
Crossref - Latif IA, AL-Azawy AF, AL-Assie AH. Assessment of genetic effects of bacterial cells after exposure to mobile phone radiation using RAPD. Iraqi J Biotechnol. 2013;12(2):63-74. https://www.iasj.net/iasj/article/79616
- AL-Azzawie AF, Jasem AS, Salih MH, Abd-albaqi MA, Sadiq ST. Evaluation of the Genetic effects of Nd: Yag and Diode lasers on Candida albicans using RAPD markers. HEZARFEN International Congress of Science, Mathematics and EngineeringAt: Izmir Ege university 2019.
- Yuan X, Song Y, Song Y, et al. Effect of Laser Irradiation on Cell Function and Its Implications in Raman Spectroscopy. Appl Environ Microbiol. 2018;84(8):e02508-17.
Crossref - Ishikawa I, Aoki A, Takasaki AA, Mizutani K, Sasaki KM, Izumi Y. Application of lasers in periodontics: true innovation or myth? Periodontol. 2000;50:90-126.
Crossref - Potenza L, Cucchiarini L, Piatti E, Angelini U, Dacha M. Effects of high static magnetic field exposure on different DNAs. Bio Electro Magnetics. 2004;25(5):352-355.
Crossref - Colbert AP, Souder J, Markov M. Static magnetic field therapy: methodological challenges to conducting clinical trials. Environmentalist. 2009;29(2):177-185.
Crossref - McLean MJ, Holcomb RR, Wamil AW, Pickett JD, Cavopol AV. Blockade of sensory neuron action potentials by a static magnetic field in the 10 mT range. Bio electro Magnetics. 1995;16(1):20-32.
Crossref - Markov MS. Therapeutic application of static magnetic fields. Environmentalist. 2007;27(4): 457-463.
Crossref - Ghodbane S, Lahbib A, Sakly M, Abdelmelek H. Bioeffects of static magnetic fields: oxidative stress, genotoxic effects, and cancer studies. BioMed Res Int. 2013;2013:1-12.
Crossref - Colbert AP, Wahbeh H, Harling N, et al. Static magnetic field therapy: a critical review of treatment parameters. Evid Based Complement Alternat Med. 2009;6(2):133-139.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.