ISSN: 0973-7510
E-ISSN: 2581-690X
Malaysia Klang River covering states of Selangor and Kuala Lumpur, has been severely polluted from urbanization, industrialization and increased hospitals. By serving as reservoir for bacterial gene transformation, Multidrug Resistant (MDR) strains especially Extended Spectrum Beta Lactamase (ESBL) resistant strains of Klebsiella pneumoniae (K. pneumoniae) have increased. To identify MDR K.pneumoniae; phenotypically determine ESBL resistant traits and molecular characterize these strains, 50 water samples were collected along Klang River and their physicochemical properties were determined. Gram-negative K. pneumoniae isolates were tested for multidrug and ESBL resistance using Kirby-Bauer method. DNA of ESBL resistant K. pneumoniae were extracted and amplified using Polymerase Chain Reaction (PCR); and conferred using blaTEM and blaCTX-M genes. Through b-lactamase gene sequencing and docking studies, the effectiveness of drugs against ESBL resistant K.pneumoniae were determined. A total of 31 (62.0%) K.pneumoniae were isolated with 12 (38.7%) positive MDR strains and 5 (41.7%) ESBL resistant strains. Only 1 (20%) blaTEM gene and 4 (80%) blaCTX-M genes were detected with Ceftriaxone (CRO) as the most effective drug showing highest binding energy (-9.2Kcal/mol) against ESBL resistant K. pneumoniae. With high prevalence of CTX-M-type ESBL resistant K.pneumoniae in Klang River, effective environmental and antibiotic controls should be adopted.
K. pneumoniae, MDR, ESBL, blaTEM, blaCTX-M.
Klebsiella pneumoniae (K.pneumoniae) from Enterobacteriaceae family is a gram negative bacteria residing in about 40% of human and animals’ intestinal tract. It is a type of normal flora localised in mouth, skin and intestine but also an opportunistic human pathogen responsible for 10% of nosocomial infections and 30-50% of exacerbations in hospitalized patients with immunosuppression. This is because K.pneumoniae not only able to colonize human skin as normal flora, it also has a persistent long survival rate in hospitals and environmental surface. Thus, transmission is relatively easy among patients in hospitals through hand contact with hospital care personnel1-3. To control the wide transmission rate and easy outbreak of K.pneumoniae, many antibiotics have been used as treatment. However, uncontrolled use of antibiotics lead to occurrence of bacterial selection pressure and caused a rise in Multidrug Resistant (MDR) strains that resist to more than one type of antibiotics4-5. MDR K.pneumoniae generally resistant to the Extended Spectrum Beta Lactamase (ESBL) antibiotics followed by Carbapenems, Aminoglycosides and Fluoroquinolones6. ESBL are plasmid mediated enzymes encoded by genes which are located mainly in the mobile genetic elements of K.pneumoniae7. According to Diagbouga and Ozturk, ESBL resistant K.pneumoniae are found to be commonly against ג-lactam antibiotics such as Penicillins, Monobactams (Aztreonam); and oxyimino Cephalosporins (Cefotaxime, Ceftazidime, Ceftriaxone, Cefuroxime, Cefepime) in which Cephalosporins are responsible for most of the ESBL antibiotic resistance in K.pneumoniae8-9. The common genes that responsible for this ESBL resistance in K. pneumoniae are CTX-M, TEM and SHV families with CTX-M b-lactamases as the most predominant and heterogeneous type5,10. The major mechanism involved in transmitting these genes is horizontal gene transfer where ESBL resistant clones are spread by sharing of plasmids across bacteria11,5,2.
The rise in resistance towards different types of antibiotics in MDR strains has become a critical issue when the resistance genes are spread into environmental bacteria especially those in highly polluted aquatic ecosystem which serve as reservoirs. Bacteria can evolves and transmits resistance among each other when in contact with human microbiota12,5. For example, resistant genes are spread into environment bacteria through discharges especially hospital waste water which contains 8 times more antibiotic resistant bacteria than domestic discharges13. The antibiotics discharged exert a selection favouring resistant bacteria by killing or inhibiting growth of susceptible bacteria. Resistant bacteria then adapt to environmental conditions and become vectors for transmission of antibiotic resistance12. Also, construction or industries effluents into aquatic ecosystem can affect the physical, chemical and biological properties of water bodies resulting in poor water quality that facilitates the evolution and transmission of resistance to environmental bacteria14. These pollutions are especially common in Klang River which covers the most densely populated areas in Malaysia including many hospitals, household areas, industries and constructions sites. The environmental degradation has gradually reduced the water quality in Klang River and the water acts as reservoir for bacterial gene transformation where environmental bacteria inter-communicates with bacteria of human or animals by horizontal gene transfer resulting in the rise of MDR strains15. Although there are many studies on the high occurrence of MDR strains in river water samples, studies on physicochemical and bacteriological status of the river are limited16-17. Thus, MDR pattern particularly ESBL resistance of K.pneumoniae in water bodies along with the physicochemical properties of Klang River are focused with TEM and CTX-M genes to be identified. Also, the most effective drug against mutated Klebsiella pneumoniae are determined.
Materials
1mol/L manganese sulphate (MnSO4), 1mol/L potassium iodide (KI) solution, 50% sulphuric acid (H2SO4), 0.018mol/L sodium thiosulphate (Na2S2O3), starch, 0.85% normal saline, Crystal violet, Gram’s iodine, Acetone, Safranin O, 50% glycerol solution.
Sample site description and collection
Malaysia Klang River covered a total of 11 major tributaries: rivers of Gombak, Batu, Kerayong, Damansara, Keruh, Kuyoh, Penchala and Kerayong, Ampang, Gisir and Kemusing with a length of 120km and drained a basin of approximately 1,288km2.18 Klang River with geographical location of 3° 13’ 01.22″, 101° 40’ 54.92″ was sourced from Klang Dam Gates located in Gombak District and ended at Port Klang, the principal port in Selangor. The upstream of Klang River in Gombak District was covered with mountains and tropical forest. The unpolluted water then flowed into the middle stream of Klang River covering the highly populated urban areas within Kuala Lumpur then towards downstream areas including the Selangor Districts of Petaling and Klang that saturated with industries and residences. From the Federal Territory of Kuala Lumpur to part of the state of Selangor in Malaysia, the river water will eventually flowed into the Straits of Malacca. The average annual temperature of Klang River was ranged between 29°C and 32°C and highest temperature was observed in March to June19-20.
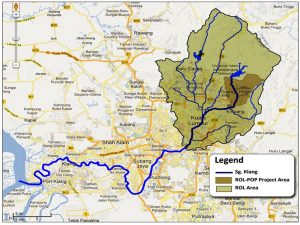
50 water samples were collected from upstream (unpolluted) to downstream (extremely polluted) of Klang River (Fig.1). Each water samples was collected along Klang River approximately 2km away on weekly basis and only when there was no rain to avoid any miscellaneous results. All samples were collected into 100ml sterile containers and 300ml dark amber bottles respectively for the estimation of Dissolved Oxygen (DO) and Biological Oxygen Demand (BOD). The bottles were tightly capped to avoid entry of air bubbles.
Fig.1. Global Positioning System (GPS) coordinates of all sampling locations along Klang River (GEC, n.d.).
Physicochemical analysis
Physicochemical parameters of water samples including physical properties, pH, temperature (0C), DO (mg/L) and BOD (mg/L) were determined on site14.
Bacterial isolation and characterization
Water samples were serially diluted by 10-folds and spread plated on MacConkey (MAC) agar. Pure isolates resembled K.pneumoniae were obtained by further streaking on Eosin Methylene Blue (EMB) agar. After incubation of plates at 37°C for 24 hours, picked colonies were transferred to Nutrient agar slants and stored in 50% glycerol solution at -80°C17,21-22.
Bacterial identification
Pure colonies resembled K.pneumoniae were inoculated in saline water and adjusted to a standard of 0.5 McFarland for Gram staining, Mannitol motility test; and biochemical tests including Indole, Methyl Red (MR), Voges-Proskauer (VP), Citrate, Urease and Triple Sugar Iron (TSI) tests 23-24.
Antibiotic resistance testing
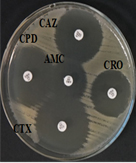
The antibiotic resistance of 31 K.pneumoniae isolated were determined using Kirby-Bauer method on Mueller Hinton Agar (MHA) according to Clinical and Laboratory Standards Institute (CLSI) standards (CLSI, 2015). Pure isolates of K.pneumoniae were inoculated in 3ml of nutrient broth and standardized at McFarland 0.5. A bacterial lawn was made on MHA. Nine b-lactam antibiotic discs including Penicillins: Amoxicillin/clavulanate (AMC), 20/10µg and Ampicillin (AMP), 10µg; second generation Cephalosporins: Cefuroxime (CXM), 30µg; third generation Cephalosporins: Cefotaxime (CTX), 30µg; Ceftazidime (CAZ), 30µg; Ceftriaxone (CRO), 30µg and Cefpodoxime (CPD), 30µg; fourth generation Cephalosporins: Cefepime (FEP), 30µg; and Monobactam: Aztreonam (ATM), 30µg were used. The plates were then inverted and incubated at 37°C for 18 hours aerobically. The zone of inhibition produced by each antibiotic discs was measured in diameter (mm) and recorded as ‘R’, ‘I’ and ‘S’ based on CLSI guidelines. Isolates with resistance to more than three types of ג-lactam antibiotics were considered as MDR strains and further confirmed with phenotypic tests25,8.
Phenotypic tests
MDR isolates equated to McFarland standard 0.5 were used to make a bacterial lawn across MHA and E.coli ATCC 25922 was used as negative control. ESBL production of K.pneumoniae was screened by Double Disc Synergy Test (DDST) using discs of AMC (20/10µg), CRO (30µg), CTX (30µg), CAZ (30µg) and CPD (30µg); and confirmed by Combination Disc Test with discs of CAZ (30µg) and Ceftazidime/Clavulanic Acid, CAZ/CA (30/10µg) according to CLSI guidelines26.
DNA extraction, PCR amplification and genotype detection
Genomic DNA from ESBL resistant K.pneumoniae were extracted using innuPREP Bacterial DNA Kit (Biometra, Germany) following manufacturer’s instructions. Extracted DNA were then amplified in a Thermal Cycler (Eppendorf, Germany) using Polymerase Chain Reaction (PCR) kit- MyTaq Mix (Bioline, United Kingdom). PCR amplification was performed using forward and reverse primers to detect TEM and CTX-M genes under specific conditions27 (Table 1). PCR products were detected using 1% gel electrophoresis and positive strains containing TEM or CTX-M gene were directed to b-lactam gene sequencing28.
Table (1):
Primers and PCR conditions for TEM and CTX-M genes detection.
blaGene |
Primers and PCR conditions |
|---|---|
TEM
|
F: 5’-ATGAGTATTCAACATTCCG-3’
R: 5’-CTGACAGTTACCAATGCTTA-3’ PCR conditions: Initial denaturation at 960C for 5 minutes; Denaturation at 960C with 35 cycles for 1 minute; Annealing at 430C for 1 minute; Extension at 720C for 1 minute; Final extension at 720C for 10 minutes; Lastly 40C forever and end. |
CTX-M |
F: 5’-ATGTGCAGYACCAGTAARGT-3’
R: 5’-TGGGTRAARTARGTSACCAGA-3’ PCR conditions: Initial denaturation at 950C for 1 minute; Denaturation with 35 cycles at 950C for 15 seconds; Annealing at 550C for 15 seconds; Extension at 720C for 10 seconds; Lastly 40C forever and end. |
Molecular docking
The forward and reverse genes’ sequence obtained were assembled into single gene sequence using BioEdit Tool then transcripted to mRNA sequence using Sequence Manipulating Suite and visualized in a 3-D structure with SWISSMODEL Server29. After determining the potential binding sites via CASTp Server and obtaining the 3-D structure of ligand (antibiotics) through PubChem database, virtual screening was performed using AutoDock-Vina to study the protein-ligand interactions with eight b-lactam antibiotics: AMP, CXM, CTX, CAZ, CRO, CPD, FEP, and ATM. A binding score (Kcal/mol) was obtained to determine drug effectiveness. The higher the binding score, the more effective was the drug30.
Physicochemical analysis
Of all 50 water samples, 31 (62.0%) water samples that were yellowish to brown, with foul smell, contained few or no debris; have a temperature of 28-32°C; and pH of 6.8-7.6 (Table 2) showed presence of K.pneumoniae. Most of the yellowish or brown, foully water samples were collected from middle and end of Klang River saturated with hospitals, housing areas, industries and animal farms. This showed that the river water was gradually contaminated from clear, odourless to yellowish or brown with fouled smell. These water samples showed a higher mean BOD (1.50±1.31mg/L; 2.54±1.94mg/L; 5.30±4.35mg/L) compared to upstream water samples that were clear or slightly clear with mean BOD (0.80±0.21mg/L; 2.95±2.04mg/L; 0.89±0.88mg/L). The highest BOD (8.85mg/L) was observed in water sample collected from end of Klang River (Port Klang) and lowest BOD (0.00mg/L) was observed in clear or slightly clear water samples collected from upstream of Klang River. When the BOD is higher compared to lower limit (2.4mg/L), the DO also showed significantly lower. The higher BOD and lower DO observed indicated the midstream, particularly end of Klang River were more polluted and have poorer water quality due to increase in total nutrients in water from discharges that encouraged growth of bacteria. Rise in amount of bacteria depleted the oxygen dissolved in water for breakdown of organic matter thus lowering DO. These results also proved the close relationship of BOD and DO, where a higher BOD will have a lower DO and vice versa. The pH of overall water samples collected (6.8-7.6) was within range of 6.5-9.0 as suggested by Gandaseca31, still feasible for aquatic life. The temperature of water samples (28-32°C) was also within the range of average annual temperature of Klang River (28-30°C) and highest temperature (32°C) observed was common as the time of sample collection was between March-June.
Table (2):
Physicochemical analysis of water samples according to their physical properties.
Physical properties |
pH |
Temperature (0C) |
DO (mg/L) |
BOD (mg/L) |
Sample size (n) |
|---|---|---|---|---|---|
Clear, odourless, no debris |
6.25±0.07 |
29.0±0.00 |
0.20±0.07 |
0.80±0.21 |
2 |
Slightly clear, odourless, no debris |
6.47±0.06 |
29.0±0.00 |
0.98±0.68 |
2.95±2.04 |
3 |
Slightly clear, odourless, few debris |
7.23±0.18 |
28.9±0.79 |
0.30±0.29 |
0.89±0.88 |
12 |
Yellowish, foul smell, no debris |
7.20±0.20 |
28.7±1.16 |
0.50±0.44 |
1.50±1.31 |
3 |
Yellowish, foul smell, few debris |
7.14±0.25 |
29.4±1.25 |
0.77±0.65 |
2.54±1.94 |
27 |
Brown, foul smell, few debris |
7.13±0.58 |
30.7±0.58 |
1.77±1.45 |
5.30±4.35 |
3 |
MDR and ESBL resistant determinants
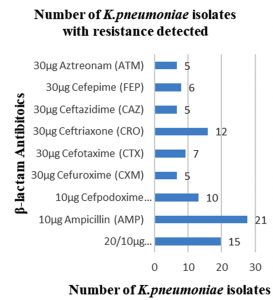
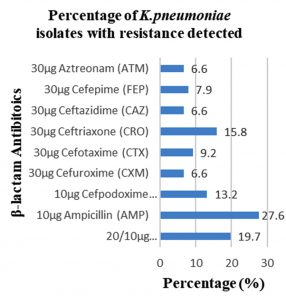
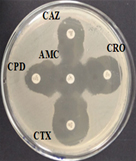
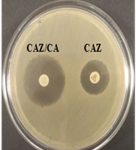
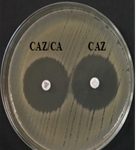
Among 31 K.pneumoniae isolated, 12 (38.7%) were MDR strains and most of the isolates were resistant to AMP showing a high prevalence of MDR K.pneumoniae (21, 27.6%) towards ג-lactam antibiotics (Fig.2 & Fig.3). Through phenotypic DDST and CDT, 5 ESBL strains (41.7%) were isolated. The ESBL production was identified from presence of ‘key-hole’ inhibition towards AMC in DDST; or a ≥5mm increase in inhibition zone of CAZ/CA compared to CAZ in CDT with reference to negative control strain E.coli ATCC 25922 (Fig.4a-d). These results confirmed K.pneumoniae was one of the most common MDR and ESBL producing bacteria to be isolated from river water samples as correlated with similar studies performed in river of Ethiopia12; in River Danube32.
Fig. 4a & 4b. DDST and CDT of K35 strain with keyhole inhibition (4a) and ≥5mm increase in zone of inhibition in CAZ/CA than CAZ alone (4b).
Agarose gel electrophoresis (AGE) and PCR amplification
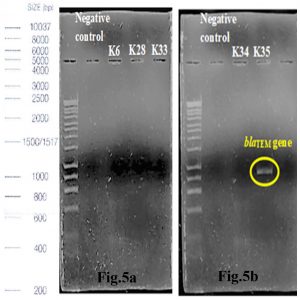
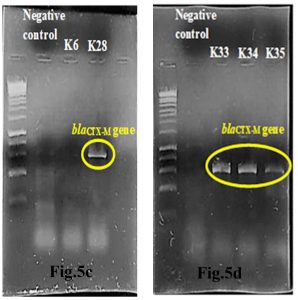
Out of 5 amplified ESBL strains, only 1 (20%) blaTEM gene and 4 (80%) blaCTX-M genes were detected through the demonstration of DNA bands of specific sizes under alpha-imager. K35 contained both TEM and CTX-M genes whereas K28, K33, and K34 contained only CTX-M genes when comparing amplicons’ bands with 1kbp standard DNA ladder to determine the size of DNA fragments (Fig.5a-d). Higher frequency of recovering CTX-M gene in this research showed that CTX-M was a predominant ESBL gene compared to other common ESBL genes such as TEM and SHV families.
Fig. 5a & 5b. Interpretation of agarose gel electrophoresis for blaTEM gene in ESBL producing K.pneumoniae with expected size of 869bp in reference to a standard 1kbp DNA ladder.
Fig.5c & 5d. Interpretation of agarose gel electrophoresis for blaCTX-M gene in ESBL producing K.pneumoniae with expected size of 869bp in reference to a standard 1kbp DNA ladder.
Molecular docking
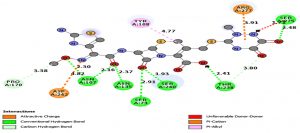
From the molecular docking study of CTX-M gene in K35, SER73, ASN107, TYR108, SER133, ASN135, PRO170, SER240, ASP242, SER275 and ARG277 (Table 3) showed to be the amino acids that were more involved in binding to eight common b-lactam antibiotics including most of the 3rd and 4th generation Cephalosporins which were the main cause of ESBL resistance in K.pneumoniae. These findings were essential particularly in designing new drugs based on existing antibiotics where functional groups in the antibiotics that allowed binding of respective amino acids can be removed, prohibiting protein-ligand interactions that encouraged enzyme hydrolysis resulting in ESBL resistance. This method is more recommended and convenient instead of changing the amino acids in mutated structure by point mutations to prohibit protein-ligand binding interactions. This is because point mutations are more challenging, time consuming and not cost effective.
Table (3):
Comparison of amino acids in CTX-M that involved in binding to ligand.
| Antibiotics | AMP | CXM | CTX | CAZ | CRO | CPD | FEP | ATM |
|---|---|---|---|---|---|---|---|---|
| Amino Acids Involved | SER 73 | SER 73 | SER 73 | SER 73 | SER 73 | TYR 108 | SER 73 | SER 73 |
| TYR 108 | ASN 107 | ASN 107 | ASN 107 | ASN 107 | PRO 170 | ASN 107 | TYR 108 | |
| SER 133 | TYR 108 | TYR 108 | ASN 135 | TYR 108 | SER 240 | TYR 108 | SER 133 | |
| GLU 274 | SER 133 | SER 133 | ASN 173 | ASN 135 | ASP 242 | SER 133 | SER 240 | |
| SER 275 | ASN 135 | ASN 135 | SER 240 | PRO 170 | ASN 135 | GLY 241 | ||
| PRO 170 | GLY 241 | ASP 242 | THR 238 | PRO 170 | SER 275 | |||
| THR 219 | ASP 242 | GLN 271 | SER 240 | LYS 237 | ||||
| LYS 237 | ALA 273 | GLU 274 | ASP 242 | SER 240 | ||||
| SER 240 | SER 275 | SER 275 | SER 275 | ASP 242 | ||||
| ASP 242 | ARG 277 | ARG 277 | ||||||
| ARG 277 |
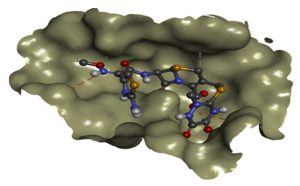
Effectiveness of drug were also determined by comparing the energy released during protein-ligand interactions (binding energy). CRO with the highest binding energy (-9.2Kcal/mol) followed by FEP (-8.7Kcal/mol), CXM (-8.4Kcal/mol), CPD and CAZ (-7.5Kcal/mol), CTX (-7.4Kcal/mol), ATM (-7.0Kcal/mol) and AMP (-6.5Kcal/mol) suggested more energy was released and weakest activity of b-lactamase with CRO (Fig.6a and Fig.6b). Thus, CRO was most unlikely to be hydrolyzed by the b-lactamase, making the drug effective.
Fig. 6b. Docking interactions of CRO with modelled blaCTX-M structure (BINDING ENERGY: -9.2 Kcal/mol).
SER 133 |
TYR 108 |
TYR 108 |
ASN 135 |
TYR 108 |
SER 240 |
TYR 108 |
SER 133 |
GLU 274 |
SER 133 |
SER 133 |
ASN 173 |
ASN 135 |
ASP 242 |
SER 133 |
SER 240 |
SER 275 |
ASN 135 |
ASN 135 |
SER 240 |
PRO 170 |
ASN 135 |
GLY 241 |
|
PRO 170 |
GLY 241 |
ASP 242 |
THR 238 |
PRO 170 |
SER 275 |
||
THR 219 |
ASP 242 |
GLN 271 |
SER 240 |
LYS 237 |
|||
LYS 237 |
ALA 273 |
GLU 274 |
ASP 242 |
SER 240 |
|||
SER 240 |
SER 275 |
SER 275 |
SER 275 |
ASP 242 |
|||
ASP 242 |
ARG 277 |
ARG 277 |
|||||
ARG 277 |
*SER=serine; TYR=tyrosine; GLU=glutamic acid; ASN=asparagine; PRO=proline; THR=threonine; LYS=lysine; ASP=aspartic acid; ARG=arginine; GLY=glycine; ALA=alanine; GLN=glutamine
In conclusion, Klang River with major pollutions has harbored high prevalence of MDR resistant (38.7%), especially ESBL resistant K.pneumoniae (41.7%) with CTX-M gene as the predominant type (80%) due to uncontrolled use of antibiotics and discharges that increase bacterial resistance by selection pressure. In order to overcome the rise of MDR strains particularly ESBL resistance in bacteria, effective waste treatment management and antibiotic control should be adapted. For example, reduce the use of AMP with replaced with CRO which is a more effective drug. From overall findings of research, the objectives were achieved and hypothesis accepted.
In future studies, modified DDST instead of general DDST method should be used to reduce risk of false negative results from interference of AmpC. In addition, more ESBL genes types should be identified to determine its prevalence rate. Besides, more molecular techniques such as Real Time PCR (qPCR) was suggested to further confirmed presence of ESBL resistance genes by identifying the exact size of DNA fragments. Location of resistance genes (plasmids, transposons or integrons) can also be determined. A wider range of antibiotics should also be used in docking studies to confirm the most effective drug.
ACKNOWLEDGEMENT
The authors wish to acknowledge the financial support by Department of Biomedical Sciences, MAHSA University.
CONFLICT OF INTERESTS
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors have made substantial, direct and intellectual contribution to the work and approved for publication.
FUNDING
None.
DATA AVAILABILITY
All data sets generated or analyzed during this study are included in manuscript.
ETHICS STATEMENT
This research work does not contain any studies related with human or animal participation.
- Pessoa-Silva, Moreira, Almeida, Flannery, Lins, Sampaio, Teixeira, Miranda, Riley and Gerberding. Extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a neonatal intensive care unit: risk factors for infection and colonization. The Journal of Hospital Infections, 2003; 53(3): 198-206.
Crossref - Rampure, Gangane, Oli, And Chandrakanth. Prevalence of MDR-ESBL producing Klebsiella pneumoniae isolated from clinical samples. Journal of Microbiology and Biotechnology Research, 2013; 3(1): 32-9.
- Wasfi, R., Elkhatib, F., and Ashour, M. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Nature Scientific Report [online], 2016; 1(6): 38929.
Crossref - Moges, Endris, Belyhun, and Worku .Isolation and characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital in northwest Ethiopia. Biomed. Central, 2014; 8(7): 215.
Crossref - Founou, L., founou, C., Allam, M., Ismail, A., Djoko, F., and Essack, Y. Genome sequencing of extended-spectrum b-lactamase (ESBL)-producing Klebsiella pneumoniae isolated from pigs and Abattoir workers in Cameroon. Frontiers in Microbiology, 2018; 9(18): 188.
Crossref - Marchaim, Navon-Venezia, Schwaber, and Carmeli. Isolation of Imipenem-resistant Enterobacter Species: emergence of KPC-2 Carbepenemase, molecular characterization, epidemiology, and outcomes. Antimicrobial Agents and Chemotherapy, 2008; 52(4): 1413-8.
Crossref - Madhavi, S., Rao, M., and Janardhan R. Bacterial etiology of acute exacerbations of chronic obstructive pulmonary disease. Journal of Microbiology and Biotechnology Research, 2012; 2(3): 440-4.
- Diagbouga, S., Salah, F., Sadji, A., Dabire, A., Nadembega, C., Kere, A., Soubeiga, S., Ouattara, A., Zohoncon, T., Belemgnegre, M., Karou, S., and Simpore, J. Detection of high prevalence of TEM/SHV/CTX-M genes in ESBL producing and multidrug resistant Klebsiella pneumoniae and Klebsiella oxytoca. Journal of Clinical Diagnosis and Research, 2016; 4(1): 130.
- Ozturk, Ozkirimli, and OzguR. Classification of beta-lactamases and penicillin binding proteins using ligand-centric network models. PLOS One, 2015; 10(2).
Crossref - Bethel, Taracila, Shyr, Thomson, Distler, Hujer, Endimiani, Wallace, Bonnet and Bonomo. Exploring the inhibition of CTX-M-9 by b-lactamase inhibitors and carbapenems. Antimicrobial Agents and Chemotherapy, 2011; 62(1): 3465-75.
Crossref - Perovic, Singh-Moodley, Duse, Bamford, Elliott, Han, Kularatne, Lowman, Whitelaw, Nana, Wadula, Lekalakala, Saif, De-Smit, and Marais. National sentinel site surveillance for antimicrobial resistance in Klebsiella pneumoniae isolates in South Africa, 2010-2012. The South African Medical Journal, 2014; 104(8): 563-8.
Crossref - Shi, W., Li, K., Ji, Y., Jiang, Q., Wang, Y., Shi, M., and Mi, Z. Carbapenem and cefoxitin resistance of Klebsiella pneumoniae strains associated with porin OmpK36 loss and DHA-1 b-lactamase production. Brazilian Journal of Microbiology, 2013; 44(2): 435-42.
Crossref - Ventola. The antibiotic resistance crisis. Journal of Pharmacy and Therapeutics, 2015; 40(4): 277-83.
- Hussain, Roohi, Munir, Ahmed, Khan, Hermann, Kim, and Anees. Biochemical characterization and identification of bacterial strains isolated from drinking water sources of Kohat, Pakistan. African Journal of Microbiology Research, 2013; 7(16): 1579-90.
Crossref - Coutinho, F., Silveira, C., Pinto, L., Salloto, G., Cardoso, A., Martins, O., Vieira, R., and Clementino, M. Antibiotic resistance is widespread in urban aquatic environments of Rio de Janeiro, Brazil. Journal of Microbial Ecology, 2014; 68(3): 441-52.
Crossref - Skariyachan, Lokesh, Rao, Kumar, Vasist, and Narayanappa. A pilot study on water pollution and characterization of multidrug resistant superbugs from Byramangala tank, Ramanagara district, Karnataka, India. Environmental Monitoring and Assessment, 2013; 185(3): 5483-95.
Crossref - Skariyachan, Mahajanakatti, Grandhi, Prasanna, Sen, Sharma, Vasist, and Narayanappa. Environmental monitoring of bacterial contamination and antibiotic resistance patterns of the fecal coliforms isolated from Cauvery River, a major drinking water source in Karnataka, India. Environmental Monitoring and Assessment, 2015; 187(5): 279-91.
Crossref - Akrami and Shafie. Improving rainfall forecasting efficiency using Modified Adaptive Neuro-Fuzzy Inference System (MANFIS). Water Resources Management, 2013, 27(9).
Crossref - Sezer, Pradhan, and Gokceoglu. Manifestation of an adaptive neuro-fuzzy model on landslide susceptibility mapping: Klang valley, Malaysia. Expert Systems with Applications, 2010; 38(7): 8208-19.
Crossref - Sharif, M., Kusin, M., Ashaari, H., and Aris, Z. Characterization of water quality conditions in the Klang River Basin, Malaysia using self organizing map and K-means algorithm. Procedia Environmental Sciences, 2015; 30(15): 73-8.
Crossref - Akoachere, T., Omam, A., and Massalla, N. Assessment of the relationship between bacteriological quality of dug-wells, hygiene behaviour and well characteristics in two cholera endemic localities in Douala, Cameroon. BMC Public Health, 2013; 13(1): 692.
Crossref - Sivamanikandan, P., and John, S. Impact of physico-chemical parameters on bacterial population in Mullaiperiyar River water-Theni district, Tamilnadu, India. African Journal of Microbiology Research, 2015; 9(1): 26-32.
Crossref - Thairu, Nasir, and Usman. Laboratory perspective of gram staining and its significance in investigations of infectious diseases. Sub-Saharan African Journal of Medicine, 2014; 1(4): 168-174.
Crossref - Talaiekhozani, Alaee, and Ponraj . Guidelines for quick application of biochemical tests to identify unknown bacteria. Accounts of Biotechnology Research, 2015; 29(2): 65-82.
- Al-Marzooq, Yusof, and Tay. Molecular analysis of antibiotic resistance determinants and plasmids in Malaysian isolates of multidrug resistant Klebsiella pneumoniae. PLOS One, 2015; 10(7): e0133654.
Crossref - Ahmed, I., El-Hady, A., Ahmed, M., and Ahmed, Z. Detection of blaSHV and blaCTX-M genes in ESBL producing Klebsiella pneumoniae isolated from Egyptian patients with suspected nosocomial infections. The Egyptian Journal of Medical Human Genetics, [online], 2013; 14(3): 277-83.
Crossref - Salah, Azab, Halaby, and Hanora. Mutations in b-lactamases detected in multidrug resistant gram negative bacteria isolated from community acquired urinary tract infections in Assiut, Egypt. African Journal of Microbiology Research, 2016; 10(46): 1938-43.
Crossref - Du, D., Wang, Z., Nathan, R., Jarrot, E., Klimont, E., Ohene-Agyei, T., Venter, H., Chiu, W., and Luisi, F. Structure of the AcrAB-TolC multidrug efflux pump. Nature, 2014; 509: 512-5.
Crossref - Thiyagarajan, C., Shanthi, S., Karthikeyan, M., and Thiruneelakandan, G. Molecular modelling and docking studies of Murb inhibitors for Klebsiella pneumoniae infections. Drug Invention Today, 2014; 4(1): 46-52.
- Chetia, H., Sharma, K., Sarma, R., and Verma, A. An in silico approach to discover potential inhibitors against multi-drug resistant bacteria producing New-Delhi metallo-b-Lactamase 1 (NDM-1) enzyme. International Journal of Pharmacy and Pharmaceutical Sciences, 2014; 6(4): 299 – 303.
- Gandaseca, S., Rosli, N., Ngayop, J., and Arianto, I. Status of water quality based on the physicochemical assessment on river water at wildlife sanctuary Sibuti Mangrove forest, Miri Sarawak. American Journal of Environmental Sciences, 2011; 7(3): 269-75.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.