ISSN: 0973-7510
E-ISSN: 2581-690X
Helicobacter pylorus (H. pylori) is a deadly bacterium responsible for significant worldwide Gastric Cancer (GC) related mortality. The present study aimed to screen all the anti-microbial drugs used to eradicate H .pylori infection and to identify the most efficient drug by using computational methods through molecular docking analysis. The 3-D structure of protein chorismate synthase of H. pylori was downloaded from the Protein data bank (PDB) online browser. The x-ray crystallography structures of 13 common drugs used against H.pylori infection were also downloaded from the drug bank. We screened all 13 common drugs through molecular docking to know the most efficient binding interaction between the diverse ligand-protein complexes. The results were further compared with clinical survey data from the patients with diverse gastrointestinal H. pylori infected cases. Among the screened compounds, by in-silico approach we found that fluoroquinolone (FLRQ) and tetracycline (TET) manifested more significant interactions with chorismate synthase (CS) protein along with binding energies of -9.2 and -8.1 kcal/mole respectively. Further, the drugs were also corroborated with the survey data from patients with varied gastrointestinal disorders in our study. With this computational study, we could find FLRQ and TET may be the most efficient drug for H. pylori treatment, which can be tried in case of anti H. Pylori treatment failure due to resistance. Hence, effective inter-analysis between the experimental and computational approaches is crucial to build up a strong inhibitor.
Antibiotics, Auto-dock, Chorismate synthase (CS), Helicobacter pylori (H. pylori)
Helicobacter pylori (H. pylori) is a bacterium known to cause chronic gastroduodenal inflammation, which can eventually lead to peptic ulcer disease (PUD) and gastric cancer (GC).1 Almost half of global population are chronically infected with H. pylori with varying degree of symptoms; asymptomatic gastritis which can lead to PUD and GC in long run.2 World Health Organization (WHO) and the International Agency for Research on Cancer (IARC) has classified H. pylori bacteria as class I carcinogen since 1994.3 The conventional eradication treatment is done with the combination of three molecules in such a way that two of them are antimicrobial agents including amoxicillin, clarithromycin, optionally metronidazole and the third drug is a proton pump inhibitor (PPI).4,5 Nevertheless, therapeutic regimen for H. pylori infection is chosen empirically on the basis of resistance pattern of the bacterium in corresponding region.1 India being a developing country had a high prevalence of H. pylori infection particularly in rural areas but still there may be differences in prevalence in every geographic region.
The antibiotic Amoxicillin (AMX) (C16H19N3O5S) has molecular weight 365.4 g/mol and is under the drug class β-lactam antibiotic. The other antibiotic Metronidazole (MTZ) (C6H9N3O3) has molecular weight 171.15 g/mol and falls under Nitroimidazoles group of antibiotics; while Tetracycline (TET) (C22H24N2O8) has molecular weight 444.435 g/mol, which comes under the drug TET antibiotics. On the other hand, Levofloxacin (LEV) (C36H42F2N6O9) has molecular weight 740.7 g/mol, which is a third-generation fluoroquinolone (FLRQ) antibiotic and Clarithromycin (CLA) (C38H69NO13) with molecular weight 747.953 g/mol falls under the drug macrolide antibiotic.
The burning issue is that the bacterium H. pylori is resistant to most of the available conventional treatments available in the current scenario.6 More importantly, the long-term usage of antibiotics for the eradication of H. pylori reduced the specificity of many of the targeted drugs resulting in reduced therapeutic efficacy in infected patients. Furthermore, many cases of H. pylori infection are asymptomatic, which can eventually give rise to GC; making the treatment even more difficult. Such asymptomatic cases are triggering points for the development of an effective vaccine against the bacteria as an alternative therapeutic approach.1 Besides, considering vaccine development as another important paradigm, on which we need to work, we can formulate better chemotherapeutic measures against H. pylori infection. The resistance to available conventional triple drug regimen against H. pylori infection is currently increasing due to the variation in its strain among the majority of infected patients.7,8 Because of increasing pattern of bacterial resistance to conventional first line triple drug regimen, there is an alarming concern to develop a better treatment modality for H. pylori infection, for which the investigators from different corners are now trying their level best to find out the new molecules and targeted therapies.9,10
The pathogenesis of H. pylori is known and the prognosis after the infection is determined by a complex set of interactions between the host and the bacterium. Chronic active gastritis is the major disorder that occurs after H. pylori infection. Nevertheless, the severity of the disease depends upon various factors such as host factors, immune response, dietary factors, and the amount of gastric acid production in the host and the strain of bacterium.2 Host genetics plays an important role in determining the severity of the disease, particularly genetic polymorphism in the host has a significant impact on the disease acquisition and progression after infection.2 Additionally, the colonization of H. pylori leads to activation of stronger immune response, but complete clearance of the infecting bacterium does not occur most of time. In such circumstances, phagocytic cells are recruited to the site of infection to stabilize the innate immunity, where the reactive oxygen species and nitrogen species are produced.2,11 However, H. pylori disrupts nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, subsequently, superoxide anions are released into the extracellular medium with augmentation within the bacteria-containing phagosomes, which leads to host tissue damage and subsequently causes ulceration by inhibiting appropriate immune response.12,13 In India, 85% of H. pylori strains are resistant to metronidazole, whereas tetracycline has a relatively lower antibacterial resistance compared to other antibiotics.14 There is a very high chance of getting resistance to metronidazole in Odisha, as well as the nearby state West Bengal, which has a similar dietary and life style pattern like Odisha. Prevalence of antimicrobial resistance in the H. pylori-infected population of Karnataka region showed very high degree of resistance to metronidazole antibiotics among the infected people.15 Interestingly, CLA resistant strains demonstrated A2143G, G2224A, C2245T and T2289C mutations in the 23 rRNA gene, whereas LEV resistance in H. pylori is primarily acquired by amino acid substitution at GyrA protein involving N87 and D91.16
Molecular docking analysis is an in silico mode approach to study the binding potential of small molecules to the given targeted protein. Molecular docking has been created and growing since long time17, yet its capacity to produce a feasible drug is still to be addressed for any disease.
In our study we have used this technique to identify the strongest binding interaction between the most commonly used antibiotics against H. pylori infection and the targeted protein Corismate synthase (CS). CS is an important enzyme in sikimate pathway, found primarily in bacteria, fungi, and plants but absent in mammals. This makes the shikimate pathway as a specific target for anti-microbial therapies. Additionally, we have also substantiated our data with an observational drug study within a group of patient with various H. pylori related gastrointestinal symptoms. We also performed secondary data analysis with the available datasets in the department at Institute of Medical Sciences & SUM Hospital, Bhubaneswar, India.
All the computational analysis was done on Intel core i7 processor, running on windows version 10 operating system (OS). The binding interaction prediction between the drugs and receptor protein CS was done with autodock software and molecular dynamic simulation studies were performed by Gromac version 2018.4 (molecular dynamic package). The molecular docking was performed with AutoDock version 4.2.6 (Molecular modeling simulation software) and graphical analysis was carried out with pymol on Molecular Graphics System, Version 2.0 Schrodinger, LLC (2015).
Molecular docking analysis
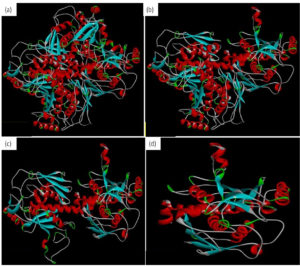
This study performed molecular docking of selected drugs that may interact with Chorismate synthase protein (1UFM) as an anti H. pylori target (Fig. 1). The 3 Dimensional (3D) protein structure of 1UFM was downloaded through online browser from protein data bank (PDB). After this, the protein 1UFM was visualized by Discovery studio visualizer. The protein 1UFM has 4 chains and all are identical. Therefore, 3 identical chains were deleted and one single chain used as a target for molecular docking (Fig. 1a, 1b, 1c). Here Fig. 1d indicates the single chain protein 1UMF.
Fig. 1. Protein 1UFM and residues. (a) With 4 chain of 1UFM. (b) With 3 chain of 1UFM. (c) With 2 chain of 1UFM. (d) With (single chain of 1UFM( used for molecular docking study).
Thirteen drugs were selected for interacting with protein 1UFM (Table 1). The drugs included in this study are Clarithromycin (CLA), Metronidazole (MTZ), Amoxicillin (AMX), Tetracycline (TET), Levofloxacin (LEV), Pantoprazole (PPZ), Tinidazole (TIND), Fluoroquinolone (FLRQ), Cisapride (CSP), Rabeprazole (RBP), Esomeprazole (EMP), Azithromycin (AZM), and Erythromycin (ETM). We carried out the survey on well-oriented prediction binding pattern, which can form a stable complex by using the conventional antibiotics and also proton pump inhibitors which are efficacious against H. pylori (Table 1).
Table (1):
List of conventional drugs used for H. pylori infections.
No |
Molecule/Drug |
Abbreviation |
Structure |
|---|---|---|---|
1 |
Clarithromycin |
CLA |
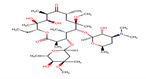 |
2 |
Metronidazole |
MTZ |
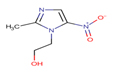 |
3 |
Amoxicillin |
AMX |
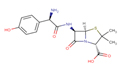 |
4 |
Tetracycline |
TET |
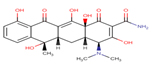 |
5 |
Levofloxacin |
LEV |
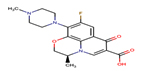 |
6 |
Pantoprazole |
PPZ |
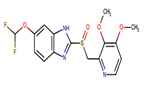 |
7 |
Tinidazole |
TIND |
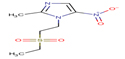 |
8 |
Fluoroquinolone |
FLRQ |
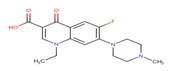 |
9 |
Cisapride; |
CSP |
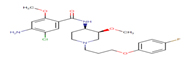 |
10 |
Rabeprazole |
RBP |
 |
11 |
Esomeprazole |
EMP |
 |
12 |
Azithromycin |
AZM |
 |
13 |
Erythromycin |
ETM |
 |
The secondary structure of the candidate drugs were drawn by the ChemDraw Ultra tool (ChembioOffice 12.0 suite). The two-dimensional structures were escalated by advanced chemistry development (ACD) labs freeware 2015, and energy was minimized by Open Babel program 2.4.1. The protein chorismate synthase found from research collaborator for structural bioinformatics (RCSB) and drug was used in discovery studio 4.5. The crystal structure of Protein-Chorismate synthase (2.25 Å) was downloaded from the PDB (accession code: 1UMF) (https://www.rcsb.org/pdb).
Molecular docking studies were employed for prediction of binding modes and free energy of designed structural molecule against targeted Chorismate synthase, through AutoDock suite program version 4.2.6. The optimal binding confirmation was shown with a semi-empirical free energy scoring function, Lamarckian genetic algorithm. Receptor-ligand inter-molecular contacts were analyzed by using BIOVIA Discovery Studio Visualizer version 4.5 and PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrodinger, LLC).18,19
Molecular dynamics (MD) Simulation and validation
Designed and synthesized conjugates were validated by MD Simulating tools. Molecular dynamics (MD) is a computational process applied for simulating the dynamic character of a molecular system as behavioral approach of the ligand protein, which simulates the dynamic behavior of the molecular systems and treats all the units in the simulating area with utmost flexibility. The principal component analysis was done to observe the motion of protein-ligand complex by performing in gmxcovar and gmxanaeig built modules of GROMACS. At the end, the binding energy was determined by using the g_mmpbsa tool.
Secondary data analysis with patients database
We planned an observational drug trial with a group of patient under treatment in our department for various H. pylori related gastrointestinal symptoms. We also assessed whether the cases with H. pylori infections were previously treated with anti H. pylori regimen, or they are denovo cases, who need to be treated by anti H. pylori regimen by the physicians. Moreover, the cases, who were already treated by the physicians of the department of Gastroenterology & Hepatobiliary Sciences, Institute of Medical Sciences and SUM Hospital, Bhubaneswar, India, their drug resistance patterns were also surveyed. The protocol was approved by Institutional Ethical Committee (Approval no.DMR/ IMS/SH/SOA/16067) and written informed consent was taken from each of the patients. All the patients under this observational study were confirmed H. pylori infected cases and were categorized on the basis of their age, sex, region, and prescribed drugs.
Statistical analysis
All the statistical analysis was performed by SPSS software version 16. The results were described as frequency/percentage as applicable.
Molecular docking study
This computational study was carried out to identify the mode of binding interaction between the drug and targeted protein. The docking study represented the perfect matching of molecular structures and the binding orientation of the drugs ( from CLA to ATM as explained) with active site of the targeted protein Chorismate Synthase of H. pylori (PDB:1UMF) as a targeted approach. Discovery studio version 4.5 program was used to perform the computational simulation. The docking scores and the amino acids formed in the H-bonding interactions and the alkyls of all the tested compounds were represented in Table 2.
Table (2):
Molecular docking score and binding interactions of commercially available H. pylori drugs with Chorismate synthase protein of H. pylori.
Compound |
Docking score (in kcal/mol) |
H-Bonding |
Alkyl |
|---|---|---|---|
CLA |
-6.9 |
Ala60, Gly92, Ser58 |
Leu235, Arg163, Lys61 |
MTZ |
-4.5 |
Gly237, His236 |
Ala167, Leu119 |
AMX |
-7.6 |
Phe156, Hist38, Gly91, Val93, Gly92 |
Val34 |
TET |
-8.1 |
Glu226 |
Lys129, Ala124, Leu119 |
LEV |
-7.3 |
Thr159 |
Leu160, Val93, His38 |
PPZ |
-7.1 |
Thr159, Gly59 |
|
TIND |
-5.6 |
Thr117, Val93, Gly92, Gly91, Thr116 |
|
FLRQ |
-9.2 |
Gly92, Thr159 |
Leu 119, Ile 33, Val 34, Leu 235 |
CSP |
-8 |
Gly237, Gly91 |
His 38, Val 34, Ile 33, Leu 235, Leu 119 |
RBP |
-7.2 |
Glu164, Val93, Gly92 |
|
EMP |
-7.5 |
Gly59 |
Val93, Leu119, Val34, Ile33 |
AZM |
-7.4 |
Tyr231, Glu164 |
Leu235, Lys61, Leu160, Val93, Leu119 |
ETM |
-7.9 |
Asp31 |
Val95, Ile94, Leu119 |
CLA, Clarithromycin; MTZ, Metronidazole; AMX, Amoxicillin; TET, Tetracycline; LEV, Levofloxacin; PPZ, Pantoprazole; TIND, Tinidazole; FLRQ, Fluoroquinolone; CSP, Cisapride; RBP, Rabeprazole; EMP, Esomeprazole; AZM, Azithromycin; ETM, Erythromycin ; Ala, Alanine; Ser, Serine; Gly, Glycine; His, Histamine; Val, Valine; Glu, Glutamic acid; Thr, Threonine; Leu, Leucine; Arg, Arginine; Lys, Lysine; Ile, Isoleucine; Tyr, Tyrosine; Asp, Aspartic acid. H-Bonding Hydrogen Bonding
Protein structure of targeted protein chorismate synthase, candidate drugs, and their binding interaction
All the 13 commonly used drugs with their abbreviation, and 2-D structures [ACD/ChemSketch having operating system Mac OS X (Microsoft windows)] were mentioned in Table 1. The 3D Structure of H. pylori receptor protein chorismate synthase was downloaded from PDB in online format (Entire Fig. 1). The interaction between these candidate drugs and Chorismate synthase protein were carried out to see protein-ligand interaction. As per our observation, docking scores of all the molecules were represented in Table 2. Hydrogen bond is one of the crucial factors influencing stability of the proteins. Our analysis demonstrated presence of both H- bonding and alkyls (Leu, Ile, Val, Leu, Lys, Ala, Leu, Leu, Arg, Lys, Ala, Leu, Val,Leu,Val,His,His,Val,Ile,Leu,Leu, Val, Leu, Val, Ile Leu, Lys, Leu, Val, Leu) during the binding interaction.
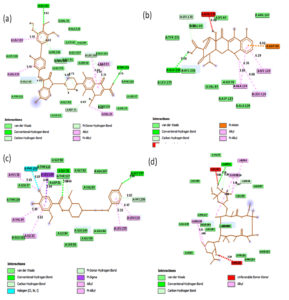
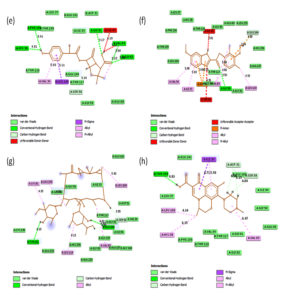
Fig. 2A. 2D interactions of compounds FLRQ, TET,CSP and ETM with chorismate synthase PDB: 1UFM. 2D structures of the candidate drugs (a) Fluoroquinolone (FLRQ). (b) Tetracycline (TET). (c) Cisapride(CSP). (d) Erythromycin (ETM).
Fig. 2B. 2D interactions of compounds AMX, EMP, AZM and LEV with chorismate synthase PDB: 1UFM. 2D structures of the candidate drugs (e) Amoxicillin (AMX). (f) Esomeprazole (EMP). (g) Azithromycin (AZM). (h) Levofloxacin (LEV).
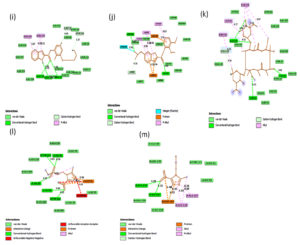
Fig. 2C. 2D interactions of compounds RBP, PPZ, CLA, TIND, and MTZ with chorismate synthase PDB: 1UFM. 2D structures of the candidate drugs (i) Rabeprazole (RBP) (j). Pantoprazole (PPZ). (k) Clarithromycin (CLA). (l) Tinadazole (TIND). (m) Metronidazole (MTZ).
Fluoroquinolone and Tetracycline showed the strongest binding interaction with Protein chorismate synthase in silico
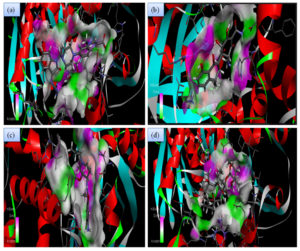
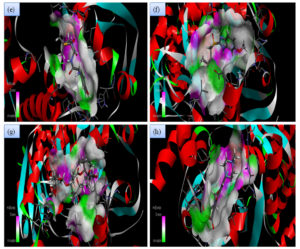
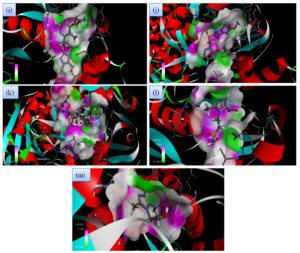
Although all the candidate drugs showed good binding interaction with the bacterial receptor chorismate synthase in silico; but we observed two most secured drug molecules such as ‘tetracycline’ and ‘fluoroquinolone’ during docking analysis. The 2-D structures of these drugs were shown in Fig. 2 with all the different sort of bonds and possible interaction points (Fig. 2a, 2b and 2c). In our study these two drugs came up with minimum binding energy and thus demonstrated strongest binding interaction with the receptor protein chorismate synthase. The 3D-interactions of FLRQ and TET with chorismate synthase of H. pylori showed the docking orientations with hydrophobic bindings, which denoted their high potential antibacterial activity (Fig. 3a and 3b). The 3D interactions of rest of the drugs with the protein were denoted in Fig. 3c, 3d, 3b and 3c.
Fig. 3A. H-Bonding interaction of compound with the active site of Chorismate synthase protein (1UMF). (a) Fluoroquinolone (FLRQ). (b) Tetracycline (TET). (c) Cisapride(CSP). (d) Erythromycin (ETM).
Fig. 3B. H-Bonding interaction of compound with the active site of Chorismate synthase protein (1UMF). (e) Amoxicillin (AMX). (f) Esomeprazole (EMP). (g) Azithromycin (AZM). (h) Levofloxacin (LEV).
Fig. 3C. H-Bonding interaction of compound with the active site of Chorismate synthase protein (1UMF). (i) Rabeprazole (RBP) (j). Pantoprazole (PPZ). (k) Clarithromycin (CLA). (l) Tinadazole (TIND). (m) Metronidazole (MTZ).
The binding energy which corresponds to drugs TET and FLRQ were: -8.1 and -9.2kcal/mole respectively, whereas other drugs showed higher binding energies (Table 2). The drug FLRQ was bonded to the cluster by amino acids (Gly 92, Thr-159) by conventional H-bonding and amino acids (Leu 119, Ile 33, Val 34, Leu 235) by alkyl bonding. TET bonded with amino acid Glu 226 by conventional H-Bonding and amino acids (Lys 129, Ala 124, Leu 119) by alkyl bonds, which were highlighted in Table 2.
Table (3):
Demographic variables of all participants.
| Range | Frequency (%) | |
|---|---|---|
| Age | 18-40 | 82(33%) |
| 41-60 | 97(40%) | |
| 61-60 | 61(25%) | |
| >81 | 5(2%) | |
| Gender | Individual | Frequency of Helicobacter positivity |
| Male | 212 | 154(73%) |
| female | 33 | 25(76%) |
| Patient Preclinical symptoms | ||
| Symptoms | Frequency (%) | |
| Upper abdominal pain | 54(22%) | |
| Bloating | 194(79% | |
| Loss of Appetite | 192(78%) | |
| Weight loss | 184(75% | |
| Clinical diagnosis | ||
| PUD | 178(73%) | |
| DU | 108(44%) | |
| Gastric ulcer | 74(30%) | |
| Gastritis | 17(7%) | |
| GERD | 13(5%) | |
| Duodenitits | 14(6%) | |
| Other non ulcer | 18(7%) | |
| Malignant growth | 28(11%) | |
| Drugs used | ||
| Apirin | 9(4%) | |
| NSAID | 29(12%) | |
| Steroids | 4(1%) | |
| Anticoagulants | 7(3%) | |
| PPI | 34(14%) | |
Clinical survey data analysis
As our results demonstrated stronger binding interaction between chorismate synthase and TET as well FLRQ, so the present study further endeavored to substantiate the results with patients’ datasets. We collected data from 245 patients and identified the most common ailment associated with H. pylori infection. We also categorized these patients as per their previous use of different types of chemotherapeutic drugs. As expected, our analysis confirmed that most of the H. pylori patients under survey were associated with peptic ulcer disease (PUD), as represented in Table 3. The survey results showed that among all the prescribed drugs, the response of TET and FLRQ was better in comparison to other drugs used, as the resistance pattern was less common with both these drugs. This result corroborated with our result, what we obtained with molecular docking analysis. As the FLRQ and TET have the susceptibility rate 86 and 67% respectively which is higher than any other drugs prescribed by the consultants and the Molecular docking scores also supported this too; therefore these two drugs might be game changer with other combination therapies. Most of our study populations were within 41-60 years of age group. Male outnumbered the female as they had more outdoor activities and therefore more prone to suffer from H. pylori infection compared to the female. Most of our patients were from Bhubaneswar, Cuttack, Puri and Nayagardh districts. Most of H. pylori infected patients had middle socio economic status. Most of the participants presented with bloating, loss of appetite, weight loss and upper abdominal pain with prevalence of 79%, 78%, 75% and 22% respectively. Most (77%) of participants were not exposed to any proton pump inhibitor (PPI) kit previously, 6% participants have used PPI Kit for 2 weeks and 5% participant has used PPI Kit for 4 weeks, as an extended regimen. Endoscopy procedure showed PUD, Gastritis, gastro esophageal reflux disorder (GERD), duodenal ulcer, Gastric ulcer, duodenitits, non ulcer dyspepsia (NUD) and malignant growth in different H. Pylori infected cases (Table 3). Additionally we also found the resistance pattern of different antibiotics surveyed in this study, which were AMX -73%, CLA -11%, MTZ- 53%, TET -47%, LEV -10%, FLRQ -13%. Finally we compared the molecular docking scores and the patients based survey data to know the correlation between the two methods highlighting the most effecting drugs during our study (Table 4).
Table (4):
Comparison of docking score and patients based survey data to represent the correlation.
Antibiotic R |
Percentage (%) |
Docking scores |
|
|---|---|---|---|
AMX |
140 |
78 |
-7.6 |
CLA |
123 |
69 |
-6.9 |
MTZ |
139 |
78 |
-4.5 |
TET |
59 |
33 |
-8.1 |
LEV |
92 |
51 |
7.3 |
FLRQ |
25 |
14 |
-9.2 |
Antibiotic R; Antibiotic Resistance, AMX, Amoxicillin; CLA, Clarithromycin, MTZ,Metronidazole; TET,Tetracycline; LEV,Levofloxacin; FLRQ,Fluoroquinolone.
H. pylori is known to be one of the most common human gastric pathogen which can cause gastric ulcer, carcinoma and duodenal ulcer, if improperly treated.20 The infection is significantly common in lower and middle socio-economic population of many developing countries. As India is a developing country, it also shows a higher prevalence of H. pylori infection and around 20 million Indians were found to be associated with PUD; which was the most common manifestation of H. pylori infection in Indians.21
Based on available literatures and our experience with the H. pylori infected patients at SUM Hospital, we explored the most commonly used antibiotics in patients of different geographical areas of Odisha. We performed in silico binding study with 13 conventional available drugs used for H. pylori eradication. The binding interaction of these drugs was studied against receptor protein chorismate synthase of H. pylori. Our results demonstrated that, the two antibiotics having highest binding affinity with the targeted protein, which might also inhibit the functions of bacterial infection and carcinogenesis, were FLRQ and TET. Chu et al22 suggested that, in addition to the reported mutation sites, H. pylori antibiotic resistance in this region may also be associated with changes in some new sites, which can lead to high recurrence rate after treatment.22,23 Similarly, in our study, FLRQ and TET revealed a promising result against H. pylori by molecular docking/in-silico study.
Salehi et al.17 during year 2019 identified key residues responsible for re-interacting GyrA with LVX, GXN and MXN and also he noticed G35 led to fully LVX resistance but the resistance pattern could be overcame by the mutation retaining all key contacts with GyrA.17 The loss of one of those contacts (i.e., G35) led to full LVX resistance. Yet, GXN and MXN overcame the mutation, by retaining all key contacts with GyrA 24. In contrast, in our study, FLRQ and TET revealed a good binding energy of -9.2 and -8.1 kcal/mole respectively against Chorismate synthase protein of H. pylori, as reported by molecular docking analysis. It is already known that FLRQ is the only drug that inhibits bacterial DNA synthesis by blocking DNA replicating enzyme gyrase and topoisomerase IV.25 Long ago it has been revealed that FLRQ are effective against H. pylori infection and the TET resistance rate is very low in adults and children i.e. 2.3 % and 2.5% respectively.26,27 Again, a meta-analysis has revealed that because of the low cost and low resistance rate, the combination of AMX and TET may be a suitable alternative treatment.28
Gehlot V et al.29 showed that more than 70% of the isolated H. pylori strains were resistant to at least one of the antibiotics used for treatment. Specially, MTZ has the highest prevalence of resistance and AMX and CLA have shown least resistance to H. pylori isolates. Hence, they suggested that these drugs might be game changer during H. pylori management in India.29 A study from Bhutan, which has high prevalence of H. pylori infection; MTZ had very high resistance rate where as CLA resistance rate was too low.30 Wani FA et al explored that TET and quinolones might be the antibiotics of choice in the eradication of H. pylori in the region of Jamu and Kashmir.23 Interestingly 85% strains from Kolkata were resistant to metronidazole and 7.5% strains were resistant to tetracycline.14
Hence, FLRQ and TET containing anti H. Pylori regimen might be the best treatment regimen for H. pylori infection eradication in India but further long term multicentre studies are required to validate our findings both in vivo and vitro in future.
Limitation
Although we could perform the computational and clinical survey analysis but could not confirm/ validate our results by Gold standard cultural analysis which is possibly the major limitation of this study.
We concluded that an effective inter-analysis in-between the experimental and computational approaches are too much crucial in the current context to find out a strong inhibitor for H. Pylori. As more and more Insilco approaches are invented, application to H. pylori drug discovery would provide novel and effective inhibitor for this emerging bacterium, which harms globally either by developing peptic ulcer, or even development of fatal gastric cancer. Our work is an endeavourer to find out an efficient targeting of protein CS in H. pylori, which proved that resistance pattern of FLRQ and TET were lowest compared to all other available anti H. pylori treatment medications, currently available.
ACKNOWLEDGMENTS
We would like to express our heartfelt thanks to President, Siksha O Anusandhan deemed to be University for providing assistance for this research work
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by the Siksha O Anusandhan deemed to be University, Bhubaneswar as Ph.D. fellowship grant number 1781611005/2017.
ETHICS STATEMENT
This study was approved by Ethics committee, IMS and SUM Hospital, Siksha O Anusandhan deemed to be University. IEC Ref No: DMR/IMS/SH/SOA/16067
AVAILABILITY OF DATA
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
- Debraekeleer A, Remaut H. Future perspective for potential Helicobacter pylori eradication therapies. Future Microbiol. 2018;13(06):671-687.
Crossref - Kusters JG, Van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19(3):449-490.
Crossref - Schistosomes IA. liver flukes and Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum. 1994,7;61:1-241. PMCID: PMC7681621
- Miftahussurur M, Yamaoka Y. Appropriate first-line regimens to combat Helicobacter pylori antibiotic resistance: an Asian perspective. Molecules. 2015;20(4):6068-6092.
Crossref - Nishizawa T, Suzuki H, Suzuki M, Takahashi M, Hibi T. Proton pump inhibitor-amoxicillin clarithromycin versus proton pump inhibitor-amoxicillin-metronidazole as first-line Helicobacter pylori eradication therapy. J Clin Biochem Nutr. 2012;51(2):114-116.
Crossref - Fagoonee S, Pellicano R. Helicobacter pylori: Molecular basis for colonization and survival in gastric environment and resistance to antibiotics. A short review. Infect Dis. 2019;51(6):399-408.
Crossref - Jadhav SG, Meshram RJ, Gond DS, Gacche RN. Inhibition of growth of Helicobacter pylori and its urease by coumarin derivatives: Molecular docking analysis. J Pharm Res. 2013;7(8):705-711.
Crossref - Bonacorsi C, Raddi MS, Carlos IZ, Sannomiya M, Vilegas W. Anti-Helicobacter pylori activity and immunostimulatory effect of extracts from Byrsonima crassa Nied. (Malpighiaceae). BMC Complement Altern Med. 2009;9(1):1-7.
Crossref - Ecclissato C, Marchioretto MA, Mendonca S, et al. Increased primary resistance to recommended antibiotics negatively affects Helicobacter pylori eradication. Helicobacter. 2002;7(1):53-59.
Crossref - Prasertpetmanee S, Mahachai V, Vilaichone RK. Improved efficacy of proton pump inhibitor-amoxicillin-clarithromycin triple therapy for Helicobacter pylori eradication in low clarithromycin resistance areas or for tailored therapy. Helicobacter. 2013;18(4):270-273.
Crossref - Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006;61(4):847-860.
Crossref - Allen LA, Beecher BR, Lynch JT, Rohner OV, Wittine LM. Helicobacter pylori disrupts NADPH oxidase targeting in human neutrophils to induce extracellular superoxide release. J Immunol. 2005;174(6):3658-3667.
Crossref - Wu Y, Antony S, Meitzler JL, Doroshow JH. Molecular mechanisms underlying chronic inflammation-associated cancers. Cancer Letters. 2014;345(2):164-173.
Crossref - Datta S, Chattopadhyay S, Patra R, et al. Most Helicobacter pylori strains of Kolkata in India are resistant to metronidazole but susceptible to other drugs commonly used for eradication and ulcer therapy. Aliment Pharmacol Ther. 2005;22(1):51-57.
Crossref - Shetty V, Lamichhane B, Tay CY, et al. High primary resistance to metronidazole and levofloxacin, and a moderate resistance to clarithromycin in Helicobacter pylori isolated from Karnataka patients. Gut Pathogens. 2019;11:21.
Crossref - Hu Y, Zhang M, Lu B, Dai J. Helicobacter pylori and antibiotic resistance, a continuing and intractable problem. Helicobacter. 2016;21(5):349-363.
Crossref - Sethi A, Joshi K, Sasikala K, Alvala M. Molecular docking in modern drug discovery: principles and recent applications. Drug Discovery and Development-New Advances. 2019;2:1-21.
Crossref - Sahoo CR, Paidesetty SK, Dehury B, Padhy RN. Molecular dynamics and computational study of Mannich-based coumarin derivatives: potent tyrosine kinase inhibitor. J Biomol Struct Dyn. 2020;38(18):5419-5428.
Crossref - Coskun GP, Djikic T, Kalayci S, Yelekci K, Sahin F, Kucukguzel SG. Synthesis, Molecular Modelling and antibacterial activity against Helicobacter pylori of novel diflunisal derivatives as urease enzyme inhibitors. Lett Drug Des Discov. 2019;16(4):392-400.
Crossref - Muzaheed. Helicobacter pylori Oncogenicity: Mechanism, Prevention, and Risk Factors. Sci World J. 2020; 2020;3018326.
Crossref - Panigrahi MK, Singh A, Uthansingh K, et al. Clinical Profile and Risk factor of Peptic ulcer disease in coastal eastern India. Journal of Medical Science And Clinical Research. 2018;6(7):615-520.
Crossref - Chu A, Wang D, Guo Q, Lv Z, Yuan Y, Gong Y. Molecular detection of H. pylori antibiotic-resistant genes and molecular docking analysis. The FASEB Journal. 2020;34(1):610-618.
Crossref - Wani FA, Bashir G, Khan MA, Zargar SA, Rasool Z, Qadri Q. Antibiotic resistance in Helicobacter pylori: a mutational analysis from a tertiary care hospital in Kashmir, India. Indian J Med Microbiol. 2018;36(2):265-272.
Crossref - Salehi N, Attaran B, Eskini N, et al. New insights into resistance of Helicobacter pylori against third-and fourth-generation fluoroquinolones: A molecular docking study of prevalent GyrA mutations. Helicobacter. 2019;24(5):e12628.
Crossref - Fontes LE, Martimbianco AL, Mochdece CC, Riera R. Proton pump inhibitor-and fluoroquinolone-based triple therapies for Helicobacter pylori eradication. Cochrane Database Syst Rev. 2017;3:1-13.
Crossref - McNulty CA, Dent JC, Ford GA, Wilkinson SP. Inhibitory antimicrobial concentrations against Campylobacter pylori in gastric mucosa. J Antimicrob Chemother. 1988;22(5):729-738.
Crossref - Peretz A, Paritsky M, Nasser O, et al. Resistance of Helicobacter pylori to tetracycline, amoxicillin, clarithromycin and metronidazole in Israeli children and adults. J Antibiot. 2014;67(8):555-557.
Crossref - Lv ZF, Wang FC, Zheng HL, et al. Meta-analysis: Is combination of tetracycline and amoxicillin suitable for Helicobacter pylori infection?. World J Gastroenterol. 2015; 21(8):2522-2533.
Crossref - Gehlot V, Mahant S, Mukhopadhyay AK, et al. Antimicrobial susceptibility profiles of Helicobacter pylori isolated from patients in North India. J Glob Antimicrob Resis. 2016;5:51-56.
Crossref - Vilaichone R-K, Aumpan N, Ratanachu-ek T, et al. Population-based Study of Helicobacter pylori infection and Antibiotic Resistance in Bhutan. Int J Infect Dis. 2020;97:102-107.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.