Leptospirosis is a widespread infectious disease caused by the spirochete Leptospira. The clinical features of leptospirosis are fever, headache, vomiting, jaundice, and the acute form of the disease is commonly called Weil’s disease. The microscopic agglutination test (MAT) is a gold standard method used to detect leptospirosis. However, it requires 14 days of time and skilled personnel to detect leptospirosis. Various molecular methods were developed for the rapid detection process, including polymerase chain reaction (PCR), multiplex PCR, nested PCR, real-time PCR, and Loop-mediated isothermal amplification (LAMP). Other immuno-based biosensor kits are readily available for the diagnosis of leptospirosis. Though these methods claim to be highly sensitive and specific, each method has its drawbacks. This review discusses the different molecular diagnostic techniques applied for the diagnosis of leptospirosis; elaborating on each method’s sensitivity, specificity, and detection time and the different samples of water, blood, and urine used.
Leptospirosis, MAT, LAMP, PCR, marker genes
Leptospirosis is a zoonotic illness caused by infection with spirochetes of the genus Leptospira. It is primarily found in tropical areas, where seasonal outbreaks are becoming more common.1 The leptospires are classified as pathogenic Leptospira and saprophytic or non-pathogenic Leptospira, and it has around 200 pathogenic serovars divided into 25 serogroups. The disease can spread to humans via direct skin contact with contaminated soil, water, and plants, as well as infected animal urine.2 In the early stage of infection, the disease shows minor symptoms such as cold, fever, headache, and jaundice; later on, it causes Weil’s illness, an acute type of leptospirosis causing liver and lung failure, resulting in death.3,4
Leptospirosis is commonly diagnosed by laboratory tests such as microscopic agglutination test (MAT) and Enzyme-linked immune sorbent assay (ELISA), which are reliable and sensitive.5 But these immunodiagnostic methods can be performed only after a week of symptomatic condition since antibodies are absent in the prodromal stage. Later, molecular diagnostics methods were developed to detect leptospirosis in the early stage of infection, even before antibodies were formed. Molecular diagnostics methods have been established with high sensitivity and specificity targeting specific genes to detect Leptospira.6 This review focuses on the various molecular methods for diagnosing leptospirosis with their advantages and disadvantages.
Leptospirosis
Leptospirosis is a life-threatening infection caused by corkscrew-shaped bacteria called Leptospira. It can be found worldwide, but it is the most prevalent in tropical and subtropical regions during late summer and after heavy rainfall.7 Leptospira can occur both as free-living or associated with a host. These species are classified into serogroups, serovars, and strains based on the antigens. Although the infection confers serovar-specific immunity, it does not necessarily prevent infection by a different serovar. The most severe manifestations of leptospirosis, which lead to death, are Weil’s disease, pulmonary haemorrhage syndrome, and hepatic failure. Although leptospirosis has a wide range of symptoms, a large proportion of infections are misdiagnosed. Its resemblance to other febrile diseases such as pneumonia, typhoid, hepatitis, and malaria complicate its diagnosis.8 Men appear to be at a higher risk than women of developing the disease. Incubation times for Leptospira vary from 2 to 21 days, with an average of ten days in humans.9 Non-pathogenic Leptospira can be found in various wet environments, such as surface water, soil, tap water, and seawater, where saprophytic halophiles are most likely to be found. In order to receive prompt and effective treatment, leptospirosis must be diagnosed as soon as possible.
Morphology
Leptospires are spirochetes, tightly coiled with hooked ends and highly motile in the longitudinal axis.8 They are ≅ 6-20 µm long and 0.1 µm in diameter. They are fragile and observed only in a phase-contrast microscope or darkfield microscope. Leptospires are obligate aerobes that can grow at optimum temperatures of 28°C to 32°C and a pH range of 6.8 to 7.4. The typical morphology, rotational movement, unique coiled-shaped body, and hooks are the structural characteristics that differentiate leptospires from other organisms.10
They have a distinctive double-membrane structure found in other spirochetes, with cytoplasmic membrane and peptidoglycan cell wall being closely associated and covered by an outer membrane. Leptospira are classified based on lipopolysaccharide (LPS), the major antigen for bacteria in the outer membrane, which shows structural heterogeneity based on gene differences.11 The genome of Leptospira is made up of two circular chromosomes, and it is more prominent when compared with genomes of other spirochetes, such as Treponema spp. and Borrelia spp., indicating the ability of Leptospira species to live in a wide range of environments.9,12
Transmission
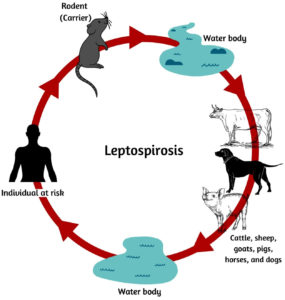
Leptospira is transmitted through contaminated water or animal urine that enters humans through open wounds or inhalation of infected samples (Fig. 1). Leptospires harbor in the kidney of animals, and temporary carriers (cattle), and permanent carriers (rodents). Endemic or epidemic episodes of leptospiral infection occur in regions with high rainfall or natural disasters.13 Humans exposed to such environments are more prone to infection. Leptospires can live in a wet environment for several months, depending on suitable environmental conditions, including soils, mud, streams, and rivers, or in organs and tissues of live or dead animals.8 The majority of mammalian species naturally carry pathogenic leptospires, which are released in the urine of infected animals, including rodents and farm animals, even though they do not show symptoms of infection. As a result, leptospirosis is a serious occupational disease that affects a wide range of people, including farmers, slaughterhouse workers, veterinarians, rodent catchers, sewer workers, and others.14,15 They can sometimes enter the human body through inhaling urine droplets or drinking water but are rarely passed from one person to another through sexual contact, transplacentally from the mother to the baby, or breast milk.16
Microbiological Diagnostic Methods
Microbiological diagnostic methods were developed to confirm the presence of Leptospira spp. in clinical samples. Some microbiological methods for detecting leptospirosis include darkfield microscopy that can be performed using direct microscopy or staining, culture-based methods and, agglutination tests.
Darkfield microscopy
Diagnosis of leptospirosis using dark field microscopy (DFM), an early diagnostic process and cost-effective method, but it has very low sensitivity, and it could not identify pathogenic organisms. Leptospires appear as thin, bright, actively motile rods that move in a rapid spinning and jerking motion. Darkfield microscopy requires approximately 10 leptospires/mL for one cell per field to be visible. Another disadvantage of this method is that it is easy to make false positive and false negative diagnoses, even by experienced hands.14 The sensitivity of DFM will decrease when the samples are collected after the first week of infection. The immunostaining method can be incorporated with darkfield microscopy to increase the sensitivity.17, 18
Culturing of Leptospira
Leptospira spp. can be isolated from blood, urine, and cerebrospinal fluid (CSF). For early detection of leptospires, blood samples from an affected person should be collected within one week of the onset of symptoms; however, after ten days of illness, leptospires in blood samples may disappear before producing any antibody and they may persist in other organs.18 Ellinghausen-McCullough-Johnson-Harris (EMJH) medium and Studdard medium are commonly used to culture leptospires.19,20 The slow growth of Leptospira in laboratories makes it a time-consuming and tedious process.17
Immunodiagnostic Methods
The mortality rate from leptospirosis has risen over time. A prompt diagnosis is required to treat this infection in its early stages. As a result, various immunodiagnostic tests are available, including microscopic agglutination tests, enzyme-linked immunosorbent assays (ELISA), and others.
Microscopic agglutination test (MAT)
The gold standard serological test for Leptospira diagnosis is MAT, which is also used as a reference test in labs for antibody detection.21 This method involves diluting a patient’s serum with live leptospires in various dilutions to test for antibodies.22 Typically 5-7 day-old culture are used as an antigen for the test. Antibodies do not appear in the patient serum until seven days after the illness, hence patient sample must be tested after the 7th day.23 The MAT is read by viewing the microtiter plate under a dark-field microscope or by placing a small drop of the reaction mixture on a microscope slide. A four-fold increase in the paired samples indicates leptospiral infection, with a titer of 1 to 100.22 Individuals with acute leptospirosis have both IgM and IgG antibodies.24 Despite its high sensitivity and specificity, the MAT takes longer to confirm positive cases. It is a complex test requiring a large panel of live-cell suspensions to adequately cover the antigenic diversity found in a given testing site. The MAT cannot be standardized because live leptospires are used as antigens.25
IgM – ELISA
The enzyme-linked immunosorbent assay (ELISA) is a serological method for detecting IgM antibodies quickly and accurately to diagnose leptospirosis. This assay can detect anti-leptospiral antibodies as early as 4-5 days after the onset of symptoms.26 It detects genus-specific antibodies, either IgM or both IgG and IgM, and is faster and more sensitive than MAT.27 The antigen used for ELISA should be obtained from leptospiral culture. However, this method is prone to producing false-negative results, which is a disadvantage.28
Marker genes and their significance
Gene markers are DNA sequences with a known physical location on a chromosome. For Leptospira detection, a variety of molecular markers are used. The lipoprotein coding genes are used as markers to identify Leptospira based on their Lipopolysaccharide (LPS), which is the main antigen in serological classification. (Table 1) lists the markers used in the molecular diagnostic method, including lipoprotein markers, outer membrane proteins (OMP), and others.
Table (1):
The marker genes targeting Leptospira with their significance.
| No | Target | Marker genes | Size (kDa) | Significance | References |
|---|---|---|---|---|---|
| 1 | Lipoprotein (Lip) | Lipl32 | 32 | Unique outer membrane protein presents only in pathogenic species. | [29, 30] |
| 2 | LipL41 | 41 | A major hydrophobic and detergent-extractable membrane protein. | [31],[32],[33] | |
| 3 | LipL36 | 36 | Present in the inner surface of the outer membrane. It plays an essential role in pathogenicity. |
[34] | |
| 4 | LipL21 | 17 | An outer membrane protein expressed on both pathogenic and non-pathogenic Leptospira. | [35] | |
| 5 | rrs | 16SrRNA | 550 | Necessary for the initiation of protein synthesis. Useful in identifying the bacteria at the species level. |
[36],[37] |
| 6 | 23SrRNA | 990 | Encoding the insertional hot spots in the L. interrogans genome | [38] | |
| 7 | Lig | ligA | 128 | Limited to L.interrogans and L. kirschneri strains | [3],[13] |
| 8 | LigB | 212 | Leptospiral immunoglobulin-like protein is widely present among pathogens and may be useful for their consistent identification and classification | [3],[13],[39] | |
| 9 | omp | ompL1 | 31 | Transmembrane is widely expressed in pathogenic leptospires. Ability to mediate attachment for various extracellular matrix (ECM) and serum components. |
[29],[40] |
| 10 | fla | flaB | 31.3 | Flagellin B protein for locomotion of the bacteria. | [32],[41],[42],[43] |
| 11 | sec | secY | 13.64 | A protein translocase subunit. It has great phylogenetic potential. |
[13],[42],[44] |
| 12 | gyr | gyrB | 71.32 | DNA gyrase B. Higher nucleotide divergence in Leptospira species |
[13] |
Samples Used For Molecular Diagnostic Methods
The stage of infection determines the clinical samples used for leptospirosis molecular diagnostics.16 Whole blood, urine, serum, post mortem samples, and cerebrospinal fluid are all examples of fluids that can be used as clinical samples for detecting Leptospira. The most common test for Leptospira is a blood sample. In the first 5 to 10 days after infection, molecular methods can be used to diagnose leptospirosis. It’s only good for two weeks’ worth of symptoms. Leptospires in the blood are gone after 10-15 days.45 Leptospira is typically transmitted from a carrier to a human through infected animals’ urine. Leptospires are found in urine for a long time and can be used to diagnose the disease at any stage.46 Organs of affected animals or humans and the aborted foetus of animals were sampled post mortem.16 The bacterial concentration in serum samples is lower than in freshly isolated blood samples, and it can be collected while the patient is still in the acute stage. A study also reported two cases of Leptospirosis wherein Leptospira nucleic acid was detected in the cerebrospinal fluid, but not in the plasma.16
Molecular diagnostic methods
Molecular methods are preferred over serological methods because they detect disease more quickly and earlier. These molecular diagnostic methods have been shown to improve detection specificity and sensitivity. Some of the molecular methods used to detect leptospirosis include polymerase chain reaction (PCR), multiplex PCR, nested PCR, real-time PCR, and loop-mediated isothermal amplification (LAMP). (Table 2) shows how these methods are designed differently depending on the sensitivity, specificity, and temperature of the sample used.
Table (2):
Molecular diagnostic methods for the detection of leptospirosis with sensitivity and specificity.
| Method | Gene target | Sample | No. of samples | Size | Sensitivity | Specificity | Reference |
|---|---|---|---|---|---|---|---|
| PCR | 23S rDNA | Urine (Dog) | 132 | 260 and 220 bp | 100% | 88.3% | Harkin et al., 2003 |
| NA | Blood and urine (Human) | 33 | 285 bp | G1/G2 – 57.6% LP1/LP2- 33.3% |
NA | Fonseca et al., 2005 | |
| 16S rRNA | Serum (Human) | 124 | NA | 62% | 100% | Fonseca et al., 2006 | |
| 16S rRNA | Serum (Human) | 100 | NA | 11% | 95% | Smita shekatkar et al., 2010 | |
| LipL32 OmpL1 16S rRNA |
Serum (Human and cattle) | 133 | 497 bp 406 bp 289 bp |
98.68% 87.01% 100% |
98.25% 100% 100% |
Gokmen et al., 2016 | |
| LipL32 | Blood (Human) | 207 | NA | 52% | 79% | Mulla summaiya et al., 2016 | |
| LipL32 | Blood (Human) | 347 | NA | NA | NA | Fernandes et al., 2019 | |
| LipL32 | Urine (Cattle) | 170 | 497bp | NA | NA | Satbige et al., 2019 | |
| Multiplex PCR | 31kDa for Brucella spp. 16S rRNA for Leptospira spp. |
Bovine fetal Sample (Hamster) |
63 | 223bp 331bp |
100% | 93% | Richtzehain et al., 2002 |
| LipL32 SecY flaB |
DNA (Water) | 32 | 423bp 285bp 793bp |
NA | NA | Moreno et al., 2010 | |
| LipL32 16SrRNA |
Water sample | 100 | 474bp 430bp |
NA | NA | Vital-Brazil et al.,2010 | |
| 16SrRNA LipL32 |
Culture DNA(Human) | 10 Leptospira spp. and 23 other | 330bp 660bp |
21.8pg | NA | Ahmed et al., 2012 | |
| 56kDA for Orienta. tsutsugamushi 17kDa for Rickettsia typhi LipL32 |
Blood (Human) | 83 | 166bp 434bp 243bp |
100% | 70% | Sea-liang et al., 2019 | |
| Nested PCR | LipL32 | Culture DNA | 35 pathogenic serovars and 7 saprophytic serovars | 183bp in 264 bp | NA | NA | Nassi et al., 2003 |
| LipL32 | Urine Serum (Human) |
59 | 183bp in 264bp | NA | NA | Jouglard et al., 2006 | |
| LipL32 | Urine (Cattle) | 30 | 859bp in 497bp | 200pg | 100% | Bomfin et al., 2007 | |
| flaB | Blood (Human) | 74 | NA | NA | NA | Aziz et al.,2019 | |
| Real-time PCR SYBR Green |
secY | Blood Serum (Human) |
133 | 245bp | 93% | 100% | Ahmed et al., 2009 |
| LipL32 16S rRNA |
Blood Serum (Human) |
266 | NA | 43% 56% |
93% 90% |
Thaipadunpanit et al., 2011 | |
| Taqman probe SYBR Green Probe based |
LipL32 16S rRNA |
Blood Urine Blood Urine (Human) |
36 | NA NA |
86% NA 100% NA |
100% 100% 97% 91.5% |
Villumsen et al., 2012 |
| secY | Blood (Human) | 111 | 203bp | 67.7% | 90% | Denipitiya et al., 2016 | |
| LipL32 | Blood, serum, urine (Human) | 119 | NA | 93% | 98.3% | Ahmed et al., 2020 | |
| Loop mediated isothermal amplification | rrs LipL41 |
Blood (Human) Blood (Human) |
133 | NA NA |
43.6% 37.6% |
83.5% 90.2% |
Sonthayanon et al., 2011 |
| rrs | Urine (Rat) | 18 | NA | 2 genome equivalent per reaction | 66.7% | Koizumi et al., 2012 | |
| 16S rRNA | DNA (Human) | 24 | 438bp | 10-100 times higher than PCR method | 100% | Suwancharoen et al., 2012 | |
| LipL32 | Serum and urine (Cat) | NA | NA | 91.69% | 100% | Hsu et al., 2017 | |
| LipL32 | Bacterial strains | 172 | 80 genome per equivalent | 200 fg/µL | 100% | Najian et al., 2019 |
Legends: NA-Not applicable
Polymerase chain reaction (PCR)
The polymerase chain reaction (PCR) is a molecular technique for amplifying a specific target DNA to a large number of copies. Since 1989, this method has become a more commonly used diagnostic method for detecting Leptospira spp. in clinical samples.46,66 PCR can be used as a complementary test in the early stages of leptospirosis, especially when no specific antibodies are found in serological reactions, allowing for an early diagnosis and separating leptospirosis from other febrile infections. The ubiquitous 16S rRNA, as well as genes for proteins like OmpL1, Lipl32, LipL36, and LipL41, have been used as the molecular target of PCR-based detection.29 Generally, the most extensively used primers for identifying leptospiral DNA target the rrs gene, encodes 16S rRNA, G1/G2-region, yields a 285bp product; flaB gene, produces a 793bp product, and B64I/B64II-region, amplify a 563bp product. Primers that target the rrs gene were unable to distinguish between pathogenic and non-pathogenic Leptospira, resulting in misdiagnosis. Nonetheless, the flaB gene was able to distinguish pathogenic from non-pathogenic leptospires.10,67 In the acute phase of illness, PCR is a sensitive and specific test with higher sensitivity than the serological method, according to some studies. The combined primer sets G1/G2, and B64-I/ B64-II, as well as cultures of blood and urine samples from 71 cases with acute Leptospiral infection, were used to compare the PCR method to the serological method. It was observed that PCR has a higher sensitivity (62%) than serology (48%). It can also be used as a confirmatory test for leptospirosis because of its high sensitivity and specificity in the diagnosis of leptospirosis.29 PCR is the preferred molecular method for leptospiral detection, but it requires time and expertise in sample preparation, as well as a minimum of 5 hours to complete. PCR is a fast, sensitive, and specific way to detect leptospiral infection, especially in the early stages of illness. When leptospires are present in significantly lower concentrations (1-10 leptospires/mL), PCR fails.68
Multiplex PCR
Multiplex PCR (mPCR) is a molecular technique that allows for simultaneous amplification of multiple primer pairs in a single tube. Several infectious diseases caused by bacteria, fungi, parasites, and viruses have been detected using this technique.56 The multiplex PCR assay is critical for developing improved health care and treatment for patients with acute indiscernible illness, especially in endemic areas.54 Several multiplex PCR techniques were used to detect pathogenic leptospirosis using primer pairs specific for the gene markers Lipl32 and 16S rRNA.54,68 With high sensitivity and specificity, this assay detected different organisms targeting different genes in a reaction. The specificity and sensitivity of the mPCR assay for sensitive and reliable detection of Salmonella, Leptospira, and Brucella spp. were tested, and the results were compared to single-PCR detection. With a limit of 100 fg for Brucella and one pg for both Salmonella and Leptospira strains, the assay appears to be highly specific and sensitive.33 Similarly, the mPCR assay detected Orientia tsutsugamushi, Rickettsia typhi, and pathogenic Leptospira spp with 100% sensitivity and 70% specificity, which was much better than serological tests.56 This method could aid in identifying causative agents during the early stages of these diseases, allowing for more timely and appropriate treatment, and the primers did not react with other bacteria. Multiplex PCR was also used in several studies to detect abortive infectious agents such as Brucella spp., Leptospira spp., and Campylobacter spp. in bovine foetal tissues. In abortive bovine samples of cattle, the mPCR showed 100 percent sensitivity and 93 percent specificity for Brucella spp. and Leptospira spp.53 The mPCR can also be a useful tool for diagnosing co-infection. While it is not a replacement for PCR, it can be used to reduce the number of tests needed and provide results more quickly and inexpensively.33 When sequencing large sequential genomic regions, the disadvantages of mPCR include the fact that using a larger number of primer pairs in a single reaction can reduce amplification efficiency and cause primer cross-reaction.
Nested PCR
Nested PCR is a PCR method that reduces non-specific binding to improve the specificity and sensitivity of a PCR amplification reaction using different pairs of secondary PCR primers. When a low copy number of targets is present in the sample, this method is typically used, and it can increase sensitivity by at least 20 folds. To amplify the Lipl32 gene using the PCR method, researchers used DNA samples from pathogenic Leptospira spp. and urine and serum samples from Leptospira spp. Both studies found that PCR had a low sensitivity when it came to amplifying the 264bp region. Another pair of primers was designed to amplify the targeting gene to increase amplification sensitivity, resulting in nested PCR amplifying 183bp region from 264bp sequence with high sensitivity.57,70 In 2008, the direct nested PCR method amplified a larger region of the sequence obtained from first-round amplification with 100 percent specificity than nested PCR followed by conventional PCR.58 Apart from the Lipl32 gene, a study recently reported that targeting the secY gene from a bovine uterine fragment using nested-PCR as an alternative to conventional PCR for non-amplifiable products yielded significant results.71 In addition, when compared to other serological methods, the nested PCR may provide 100 percent specificity. It has been shown to be a quick way to detect leptospirosis during the acute phase of infection.72 Despite the fact that nested PCR improves specificity and sensitivity, it has a high risk of contamination, which can result in false positives and negatives.57
Real-time PCR
The real-time polymerase chain reaction (RT-PCR) is a technique for quickly diagnosing infectious diseases in clinical laboratories. Real-time PCR is a rapid and sensitive alternative to traditional PCR methods such as SYBR green technology, TaqMan probes, and, more recently, Light Upon eXtension (LUX) technology for detecting pathogenic Leptospira spp. In SYBR Green and LUX-based rtPCRs, precise primer annealing is required to generate amplicon-specific fluorescence signals. In probe-based techniques, specific probe annealing is also used to increase specificity. To determine the exact melting temperature (Tm) of dye and LUX-based rtPCR,55 a further melting curve analysis is required, and the melting curve can distinguish pathogenic leptospires. The melting curve analysis without the probe is less specific than the probe method, but it can be used instead of expensive probes when amplification has been optimised.73 When comparing the sensitivity of two molecular methods, rrs real-time PCR and combination of 16S rRNA/ Lipl32 real-time PCR, it was discovered that the combination of 16S rRNA and Lipl32 has a higher sensitivity than rrs RT-PCR.74 Furthermore, RT-PCR can improve the detection limit of blood and urine samples.75 Moreover, a few other studies found that RT-PCR is more accurate than serological and microbiological methods for a variety of target genes.36 Despite the fact that RT-PCR is a more advanced method than conventional PCR, both can detect disease before antibodies are produced, which can be used as an early diagnostic method.3 In comparison to traditional PCR, quantitative real-time PCR has the advantage of quickly, accurately, and sensitively diagnosing leptospirosis. This method reduces the risk of a false-positive result while also reducing contamination.76 To interpret the results, advanced equipment with a monitor connection is required, as is training to perform the assay.
Loop-mediated isothermal amplification (LAMP)
Loop-mediated isothermal amplification77 is a nucleic acid-based amplification method that can amplify even a single copy of target DNA in samples in less than one hour under isothermal conditions. This method uses six specially developed primers, four of which are used in the LAMP’s primary stage and two of which are used in the LAMP’s later stages. LAMP is a low-cost, simple method that can be observed with either gel electrophoresis or naked eyes.55 The first application of the LAMP technique for rapid detection of leptospirosis was in 2008, when the LipL41 gene was targeted with a detection limit of 100 copies and a higher specificity.78 However, in 2011, LAMP, which used purified DNA for amplification, determined a lower detection limit of 20 copies by targeting the 16S rRNA gene.68 To make the sampling process easier, a Lepto-rrs method was developed to amplify target DNA without purification of DNA from cattle urine. This method has a higher detection limit (2 genome equivalent) than previous studies that showed heat-denatured DNA can increase sensitivity.32 LAMP targeting the Lipl32 gene in combination with the LipL41 gene was found to have a detection limit of 10 copies per reaction, indicating that Lipl32 LAMP has higher diagnostic and analytical sensitivity.30 A systematic review and meta-analysis study published in 2021 compared LAMP and PCR and concluded that LAMP has higher accuracy and sensitivity.79 Real-time LAMP, multiplex LAMP, and LigB-LAMP were among the LAMP-based methods used to diagnose Leptospira spp. Targeting Lipl32 led to the development of the real-time LAMP (RealAmp) method for high sensitivity and rapid detection of leptospirosis, and the LipL41 gene can distinguish pathogenic and non-pathogenic Leptospira spp. from environmental water samples. Within 70 minutes of reaction amplification, real-time amp has been shown to have higher sensitivity than real-time PCR.35 Multiplex PCR with lateral flow dipstick, on the other hand, has developed with 100 percent specificity.80 Another LAMP-based method, LigB-LAMP, was created to target the LigB gene using Hydroxyl naphthol blue (HNB) and SYBR green dye, which has higher specificity than the traditional LAMP method.39 All of these LAMP techniques, on the other hand, can amplify the DNA target for Leptospira spp. diagnosis with greater efficiency, sensitivity, specificity, and rapidity at a lower cost. This method does not necessitate complex equipment; the reaction can be carried out in a water bath at the proper temperature. The method is widely applicable for detecting leptospirosis due to its simplicity.
Advanced molecular method for the detection of leptospirosis
Due to their improved sensitivity and specificity, biosensor-based technology has become an excellent platform in leptospirosis diagnostics in recent years, overcoming the drawbacks of PCR-based molecular techniques. Nanoparticle-based biosensors, aptamer-based biosensors, DNA biosensors, and other biosensors are all used to detect Leptospira. One of the DNA-based biosensors designed for the detection of leptospirosis is the electrochemical DNA biosensor. It can target organisms and detects the presence of L. interrogans in samples with extreme sensitivity, specificity, and speed. AuNPs (Silver Nanoparticles) – modified multiwalled carbon nanotubes immobilised with an amino-labeled ssDNA with polyamidoamine – and graphene quantum dots with cysteine as a linker make up the biosensor,81,2. In addition, multiple cross-displacement amplification (MCDA) is a method for amplifying nucleic acids in an isothermal environment to detect more bacteria, and a label-based lateral flow dipstick biosensor (LFB) assay was developed to detect Leptospira based on antibody binding.82 MCDA and LFB were combined in a study to detect Leptospira spp. from a pure culture by targeting the LipL41 gene. All pathogenic L.interrogans were found to be positive, while all non-pathogens were found to be negative.83 Similarly, a study using a lateral flow dipstick biosensor assay combined with multiplex LAMP found that thirteen pathogenic Leptospira species, two intermediate Leptospira species, one non-pathogenic Leptospira species, and twenty-eight other bacterial species had 100% specificity.65
Future aspects of molecular diagnostic methods
Innovative diagnostic techniques could help detect leptospirosis even more in the future. Isothermal amplification and real-time LAMP are two new detection methods that have high sensitivity and specificity while also being quick. Furthermore, next-generation sequencing (NGS) has been used to diagnose leptospirosis, which may lead to a better understanding of the molecular pathogenesis and virulence evolution of Leptospira spp.85 Furthermore, once multiplex real-time PCR, microarrays, and next-generation sequencing (NGS) have been shown to be a cost-effective diagnostic tool for leptospirosis diagnosis, they may become commercially available and more widely used.86
Although MAT and ELISA are commonly used serological diagnostic methods for detecting Leptospira spp., they have several drawbacks, including the need for expert handling and a long detection time. Despite this, several molecular diagnostic methods for rapid detection have been developed, which overcome the limitations of serological methods. Depending on the molecular marker genes and clinical samples used for detection, these various molecular methods can detect the presence of Leptospiral at an earlier stage with high sensitivity and specificity in a short time. The sensitivity and specificity of several molecular techniques for diagnosing leptospirosis that have been reported so far, as well as their pros and cons, are reviewed, analysed, and compared in this study. Further studies are required for the investigation of the clinical and epidemiological implications of leptospirosis molecular detection.
ACKNOWLEDGMENTS
The authors express their gratitude to SRM Institute of Science and Technology, Chennai, Tamil Nadu, India, for their support in completing the study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Ahmad SN, Shah S, Ahmad FH. Laboratory diagnosis of leptospirosis. J Postgrad Res. 2005;51(3):195-200. PMID: 16333192
- Verma V, Goyal M, Kala D, et al. Recent advances in the diagnosis of leptospirosis. Front Biosci. 2020;25:1655-1681.
Crossref - Palaniappan RUM, Chang YF, Chang CF, et al. Evaluation of lig-based conventional and real time PCR for the detection of pathogenic leptospires. Mol Cell Probes. 2005;19(2):111-117.
Crossref - Ahmed A, Grobusch MP, Klatser PR, Hartskeerl RA. Molecular approaches in the detection and characterization of Leptospira. J Bacteriol Parasitol. 2012;3(2):1000133.
Crossref - Musso D, La Scola B. Laboratory diagnosis of leptospirosis: a challenge. J Microbiol Immunol Infect. 2013;46(4):245-252.
Crossref - Di Azevedo MI, Lilenbaum W. An overview on the molecular diagnosis of animal leptospirosis. Lett Appl Microbiol. 2021;72(5):496-508.
Crossref - Haake DA, Levett PN. Leptospirosis in humans. Leptospira and leptospirosis. 2015;387:65-97.
Crossref - Al-orry W, Arahou M, Hassikou R, et al. Leptospirosis: transmission, diagnosis and prevention. Int J Innov Stud. 2016 ;15:457.
- Tilahun Z, Reta D, Simenew K. Global epidemiological overview of leptospirosis. Int J Microbiol Res. 2013;4:9-5.
Crossref - Schreier S, Doungchawee G, Chadsuthi S, Triampo D, Triampo W. Leptospirosis: current situation and trends of specific laboratory tests. Expert Rev Clin Immunol. 2013;9(3):263-280.
Crossref - Adler B, de la Pena Moctezuma A. Leptospira and leptospirosis. Vet Microbiol. 2010;140(3-4):287-296.
Crossref - Bharathi MV, Kandavel E, Nedunchelliyan S, Muralimanohar B, Kumanan K. Prevalence of Toxoplasma antibodies by using modified direct agglutination test in dogs in Chennai. J Vet Parasitol. 2011;25:162-164.
- Toyokawa T, Ohnishi M, Koizumi N. Diagnosis of acute leptospirosis. Expert Rev Anti-Infect Ther. 2011;9(1):111-121.
Crossref - Budihal SV, Perwez K. Leptospirosis diagnosis: competancy of various laboratory tests. J Clin Diagnostic Res. 2014;8(1):199-202.
Crossref - Costa F, Hagan JE, Calcagno J, et al. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis. 2015;9(9):0003898.
Crossref - Terpstra WJ. Human leptospirosis: guidance for diagnosis, surveillance and control. World Health Organization. 2003.
- Sharma KK, Kalawat U. Early diagnosis of leptospirosis by conventional methods: one-year prospective study. Indian J Pathol Microbiol. 2008;51(2):209-211.
Crossref - Jaiswal NK, Chandrasekaran S, Padmavathy BK. Dark field microscopy an important conventional technique for the early diagnosis of leptospirosis. Int J Curr Microbiol App Sci. 2015;4:718-722. https://www.ijcmas.com/vol-4-6/N.%20K.%20Jaiswal,%20et%20al.pdf
- Bal AE, Gravekamp C, Hartskeerl RA, De Meza-Brewster J, Korver H, Terpstra WJ. Detection of leptospires in urine by PCR for early diagnosis of leptospirosis. J Clin Microbiol. 1994;32(8):1894-1898.
Crossref - Zuerner RL. Laboratory maintenance of pathogenic Leptospira. Curr Protoc Microbiol. 2006;1:12.
Crossref - Bajani MD, Ashford DA, Bragg SL, et al. Evaluation of four commercially available rapid serologic tests for diagnosis of leptospirosis. J Clin Microbiol. 2003;41(2):803-809.
Crossref - Shekatkar SB, Harish BN, Menezes GA, Parija SC. Clinical and serological evaluation of Leptospirosis in Puducherry, India. J Infect Dev Ctries. 2010;4(3):139-43.
Crossref - Safiulah SA, Saleh AA, Munwar S. Laboratory methods for diagnosing leptospirosis: a review. Bangladesh. J Med Microbial. 2009;3(1):39-43.
Crossref - Kanagavel M, Shanmughapriya S, Aishwarya KV, et al. Peptide specific monoclonal antibodies of Leptospiral LigA for acute diagnosis of leptospirosis. Sci Rep. 2017;7(1):3250.
Crossref - Cumberland P, Everard CO, Levett PN. Assessment of the efficacy of an IgM-elisa and microscopic agglutination test (MAT) in the diagnosis of acute leptospirosis. Am J Trop Med Hyg. 1999;61(5):731-734.
Crossref - Rosa MI, Reis MF, Simon C, et al. IgM ELISA for leptospirosis diagnosis: a systematic review and meta-analysis. Cien Saude Colet. 2017;22(12):4001-12.
Crossref - Picardeau M, Bertherat E, Jancloes M, Skouloudis AN, Durski K, Hartskeerl RA. Rapid tests for diagnosis of leptospirosis: current tools and emerging technologies. Diagn Microbiol Infect Dis. 2014;78(1):1-8.
Crossref - Dreyfus A, Ruf MT, Mayer-Scholl A, et al. Exposure to Leptospira spp. and Associated Risk Factors in the Human, Cattle and Dog Populations in Bhutan. Pathogens. 2021;10(3):308.
Crossref - Gokmen TG, Soyal A, Kalayci Y, Onlen C, Koksal F. Comparison of 16S rRNA-PCR-RFLP, Lipl32-PCR and OmpL1-PCR methods in the diagnosis of leptospirosis. Rev Inst Med Trop Sao Paulo. 2016;58:64.
Crossref - Hsu YH, Chou SJ, Chang CC, et al. Development and validation of a new loop-mediated isothermal amplification for detection of pathogenic Leptospira species in clinical materials. J. Microbiol. Methods. 2017;141:55-59.
Crossref - Lin XA, Chen Y, Lu Y, Yan J, Yan J. Application of a loop-mediated isothermal amplification method for the detection of pathogenic Leptospira. Diagn Microbiol Infect Dis. 2009;63(3):237-242.
Crossref - Koizumi N, Nakajima C, Harunari T, et al. A new loop-mediated isothermal amplification method for rapid, simple, and sensitive detection of Leptospira spp. in urine. J Clin Microbiol. 2012;50(6):2072-2074.
Crossref - Truong QL, Yoon BI, Hahn TW. Development of a multiplex PCR to identify Salmonella, Leptospira and Brucella species in tissue samples. Korean J Vet Res. 2012;52:75-82.
Crossref - Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol. 2009;7(10):736-747.
Crossref - Monica NI, Rathinasabapathi P, Ramya M. Development of real-time loop-mediated isothermal amplification (RealAmp) method for sensitive and rapid detection of pathogenic and non-pathogenic Leptospira. Lett Appl Microbiol. 2019;68(2):196-203.
Crossref - Thaipadunpanit J, Chierakul W, Wuthiekanun V, et al. Diagnostic accuracy of real-time PCR assays targeting 16S rRNA and Lipl32 genes for human leptospirosis in Thailand: a case-control study. PLoS One. 2011;6(1):e16236.
Crossref - Jay ZJ, Inskeep WP. The distribution, diversity, and importance of 16S rRNA gene introns in the order Thermoproteales. Biol Direct. 2015;10(35):1-10.
Crossref - Cilia G, Bertelloni F, Mignone W, et al. Molecular detection of Leptospira spp. in wild boar (Sus scrofa) hunted in Liguria region (Italy). Comp Immunol Microbiol Infect Dis. 2020;68:101410.
Crossref - Ali SA, Kaur G, Boby N, et al. Rapid and visual detection of Leptospira in urine by LigB-LAMP assay with pre-addition of dye. Mol Cell Probes. 2017;36:29-35.
Crossref - Fernandes LG, Vieira ML, Kirchgatter K, et al. OmpL1 is an extracellular matrix-and plasminogen-interacting protein of Leptospira spp. Infect Immun. 2012;80(10):3679-3692.
Crossref - Kawabata H, Dancel LA, Villanueva SY, et al. flaB Polymerase Chain Reaction (flaB PCR) and Its Restriction Fragment Length Polymorphism (RFLP) Analysis Are an Efficient Tool for Detection and Identification of Leptospira Spp. Microbiol Immunol. 2001;45(6):491-496.
Crossref - Moreno N, Agudelo-Florez P. Application of conventional and multiplex PCR assays for identification of isolates of Leptospira spp. in Colombia. Rev Peru Med Exp Salud Publica. 2010;27(4):548-556.
Crossref - Stoddard RA. Detection of pathogenic Leptospira spp. through real-time PCR (qPCR) targeting the Lipl32 gene. InPCR Microb Pathog. 2013;943:257-266.
Crossref - Ahmed A, Engelberts MF, Boer KR, Ahmed N, Hartskeerl RA. Development and validation of a real-time PCR for detection of pathogenic Leptospira species in clinical materials. PloS one. 2009;4(9):e7093.
Crossref - Blanchard S, Cariou C, Bouvet J, et al. Quantitative real-time PCR assays for the detection of pathogenic Leptospira species in urine, and blood samples in canine vaccine clinical studies: a rapid alternative to classical culture methods. J Clin Microbiol. 2021;59(7):e:0300620.
Crossref - Waggoner JJ, Pinsky BA. Molecular diagnostics for human leptospirosis. Curr Opin Infect Dis. 2016;29(5):440-445.
Crossref - Agampodi SB, Matthias MA, Moreno AC, Vinetz JM. Utility of quantitative polymerase chain reaction in leptospirosis diagnosis: association of level of leptospiremia and clinical manifestations in Sri Lanka. Clin Infect Dis. 2019;54(9):1249-1255.
Crossref - Harkin KR, Roshto YM, Sullivan JT. Clinical application of a polymerase chain reaction assay for diagnosis of leptospirosis in dogs. J Am Vet Med Assoc. 2003;222(9):1224-1229.
Crossref - de Abreu Fonseca C, Teixeira de Freitas VL, Calo Romero E, et al. Polymerase chain reaction in comparison with serological tests for early diagnosis of human leptospirosis. Trop Med Int Health. 2006;11(11):1699-1707.
Crossref - Shekatkar S, Harish BN, Parija SC. Diagnosis of leptospirosis by polymerase chain reaction. Int J Pharma Bio Sci. 2010;1(3):1-6.
Crossref - Mullan S, Panwala TH. Polymerase chain reaction: an important tool for early diagnosis of leptospirosis cases. J Clin Diagnostic Res. 2016;10(12):DC08-DC11.
Crossref - Satbige AS, Patil NA, Awati B, Sandeep H. Detection of leptospirosis in the urine of cattle in North Karnataka, South India. J Entomol Zool Stud. 2020;8(1):727-729. https://www.entomoljournal.com/archives/2020/vol8issue1/PartL/8-1-138-324
- Richtzenhain LJ, Cortez A, Heinemann MB, et al. A multiplex PCR for the detection of Brucella spp. and Leptospira spp. DNA from aborted bovine fetuses. Vet Microbiol. 2002;87(2):139-147.
Crossref - Vital-Brazil JM, Balassiano IT, Oliveira FS, et al. Multiplex PCR-based detection of Leptospira in environmental water samples obtained from a slum settlement. Mem Inst Oswaldo Cruz. 2010;105(3):353-355.
Crossref - Ahmed SA, Sandai DA, Musa S, et al. Rapid diagnosis of leptospirosis by multiplex PCR. Malays J Med Sci. 2012;19(3):9-16. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3629659/
- Sea-Liang N, Sereemaspun A, Patarakul K, et al. Development of multiplex PCR for neglected infectious diseases. PLOS Negl Trop Dis. 2019;13(7):e0007440.
Crossref - Nassi F, Seixas FK, Jouglard SD, et al. Leptospirosis diagnosis using nested-PCR. Braz J Microbiol. 2003;34:90-92.
Crossref - Bomfim MR, Barbosa-Stancioli EF, Koury MC. Detection of pathogenic leptospires in urine from naturally infected cattle by nested PCR. Vet J. 2008;178(2):251-256.
Crossref - Aziz MA, Aung MS, Paul SK, et al. First molecular identification of two Leptospira species (Leptospira interrogans and Leptospira wolffii) in Bangladesh. New Microbes New Infect. 2019;31:100570.
Crossref - Villumsen S, Pedersen R, Borre MB, et al. Novel TaqMan® PCR for detection of Leptospira species in urine and blood: pit-falls of in silico validation. J Microbiol Methods. 2012;91(1):184-190.
Crossref - Denipitiya DT, Chandrasekharan NV, Abeyewickreme W, et al. Application of a real time Polymerase Chain Reaction (PCR) assay for the early diagnosis of human leptospirosis in Sri Lanka. Biologicals. 2016;44(6):497-502.
Crossref - Ahmed AA, Goris MG, Meijer MC. Development of Lipl32 real-time PCR combined with an internal and extraction control for pathogenic Leptospira detection. Plos one. 2020;15(11):e0241584.
Crossref - Sonthayanon P, Chierakul W, Wuthiekanun V, et al. Accuracy of loop-mediated isothermal amplification for diagnosis of human leptospirosis in Thailand. Am J Trop Med Hyg. 2011;84(4):614-620.
Crossref - Suwancharoen D, Kulchim C, Chirathaworn C, Yoshida S. Development of a novel primer combination to detect pathogenic Leptospira by loop-mediated isothermal amplification. J Microbiol Methods. 2012;91(1):171-173.
Crossref - Najian AN, Foo PC, Ismail N, Kim-Fatt L, Yean CY. Probe-specific loop-mediated isothermal amplification magnetogenosensor assay for rapid and specific detection of pathogenic Leptospira. Mol Cell Probes. 2019;44:63-68.
Crossref - Gravekamp C, Van de Kemp H, Franzen M, et al. Detection of seven species of pathogenic leptospires by PCR using two sets of primers. Microbiology. 1993;139(8):1691-700.
Crossref - Natarajaseenivasan K, Vijayachari P, Sharma S, et al. FlaB PCR-based identification of pathogenic Leptospiral isolates. J Microbiol Immunol Infect. 2010;43(1):62-69.
Crossref - Lucchesi P, Arroyo GH, Etcheverria AI, Parma AE, Seijo AC. Recommendations for the detection of Leptospira in urine by PCR. Rev Soc Bras Med Trop. 2004;37(2):131-134.
Crossref - Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the Lipl32 gene. Diagn Microbiol Infect Dis. 2009;64(3):247-255.
Crossref - Jouglard SDD, Simionatto S, Seixas FK, Nassi FL, Dellagostin OA. Nested polymerase chain reaction for detection of pathogenic leptospires. Can J Microbiol. 2006;52(8):747-752.
Crossref - Di Azevedo MI, Pires BC, Libonati H, et al. Extra-renal bovine leptospirosis: Molecular characterization of the Leptospira interrogans Sejroe serogroup on the uterus of non-pregnant cows. Vet Microbiol. 2020;250:108869.
Crossref - Natarajaseenivasan K, Raja V, Narayanan R. Rapid diagnosis of leptospirosis in patients with different clinical manifestations by 16S rRNA gene based nested PCR. Saudi J Biol Sci. 2012;19(2):151-155.
Crossref - Naze F, Desvars A, Picardeau M, Bourhy P, Michault A. Use of a New High Resolution Melting Method for Genotyping Pathogenic Leptospira spp. PLoS One. 2015;10(7):e0127430.
Crossref - Wood PL, Steinman M, Erol E, Carter C, Christmann U, Verma A. Lipidomic analysis of immune activation in equine leptospirosis and Leptospira-vaccinated horses. Plos one. 2018;13(2):e0193424.
Crossref - Karuniawati A, Yasmon A, Ningsih I. Optimizing real-time PCR method to detect Leptospira spp. in human blood and urine specimens. Med J Indones. 2012;21(1):13-17.
Crossref - Merien F, Portnoi D, Bourhy P, Charavay F, Berlioz-Arthaud A, Baranton G. A rapid and quantitative method for the detection of Leptospira species in human leptospirosis. FEMS Microbiol Lett. 2005;249(1):139-147.
Crossref - Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):63.
Crossref - Xu J, Xiong Y, Chen S, Guan Y. A lamp light-emitting diode-induced fluorescence detector for capillary electrophoresis. Talanta. 2008;76(2):369-372.
Crossref - Gunasegar S, Neela VK. Evaluation of diagnostic accuracy of loop-mediated isothermal amplification method (LAMP) compared with polymerase chain reaction (PCR) for Leptospira spp. in clinical samples: A systematic review and meta-analysis. Diagn Microbiol Infect Dis. 2021;100(3):115369.
Crossref - Najian ABN, Syafirah EAREN, Ismail N, Mohamed M, Yean CY. Development of multiplex loop mediated isothermal amplification (m-LAMP) label-based gold nanoparticles lateral flow dipstick biosensor for detection of pathogenic Leptospira. Anal Chim Acta. 2016;903:142-148.
Crossref - Nagraik R, Sethi S, Sharma A, Kumar D, Kumar D, Kumar AP. Ultrasensitive nanohybrid electrochemical sensor to detect Lipl32 gene of Leptospira interrogans. Chem Pap.-Slovak Acad Sci. 2021;75(10):5453-5462.
Crossref - Niu L, Zhao F, Chen J, et al. Isothermal amplification and rapid detection of Klebsiella pneumoniae based on the multiple cross displacement amplification (MCDA) and gold nanoparticle lateral flow biosensor (LFB). Plos one. 2018;13(10):e0204332.
Crossref - Li S, Liu Y, Chen X, Wang M, Hu W, Yan J. Visual and rapid detection of Leptospira interrogans using multiple cross-displacement amplification coupled with nanoparticle-based lateral flow biosensor. Vector Borne and Zoonotic Dis. 2019;19(8):604-612.
Crossref - Vishwakarma A, Lal R, Ramya M. Aptamer-based approaches for the detection of waterborne pathogens. Int Microbiol. 2021;24(2):125-140.
Crossref - Brehm TT, Zur Wiesch JS, Lutgehetmann M, et al. Epidemiology, clinical and laboratory features of 24 consecutive cases of leptospirosis at a German infectious disease center. Infect. 2018;46(6):847-853.
Crossref - Techawiwattanaboon T, Patarakul K. Update on molecular diagnosis of human leptospirosis. Asian Biomed. 2019;13(6):207-216.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.