ISSN: 0973-7510
E-ISSN: 2581-690X
The present study aimed at investigating the spread of serine carbapenemase-producing isolates of Pseudomonas aeruginosa recovered from some hospitals and specialized health centers in Al-Diwaniyah City, Iraq. Using morphological and molecular methods, 630 clinical samples (244 burn swabs, 163 sputa, 115 urine samples, and 108 ear swabs) were tested. The current work also explored the carbapenemase production ability of the carbapenemresistant (CR) isolates using morphological and molecular methods. A monoplex polymerase chain reaction (MPCR) technique targeting 4 different serine carbapenemase (SCMs) coding genes was followed. The study also explored the nucleotide sequences of the SCM genes. The API 20 E and Vitek 2 system showed that 100 of the isolates belonged to P. aeruginosa. With different severity of resistance by percentages of isolates to various antibiotics, only 24% of the isolates displayed resistance against carbapenem. The results of the SCM-related MPCR showed the occurrence of the blaGES, blasMe, blaKPC, and blaOXA-48 genes in 20 (83.3%), 19 (79.1%), 11 (45.8%), 18 (75%), respectively, of the isolates. The phylogenetic study analysis revealed high, 99%, identity with NCBI-related world strains. According to the best of our knowledge, this is the first study in Iraq that detects a group of encoding genes of serine carbapenemase in carbapenem-resistant isolates.
PCR, phylogeny, Pseudomonas aeruginosa, sequencing, serine carbapenemase genes.
Pseudomonas aeruginosa is one of the main opportunistic pathogens, responsible for nosocomial infection and infections in cancer patients undergoing chemotherapy, cystic fibrosis, and patients with severe burns. These bacteria are linked to high morbidity and mortality rates (Schaber et al., 2007; Alikhani et al., 2017; Maroui et al., 2017)The emergence of P. aeruginosa strains with multi-drug resistance (MDR) due to their possession of huge genetic modification that can develop a large number of factors associated with resistance to different antibiotics (Bassetti et al., 2018). The bacterial resistance to antibiotics is thus considered as one of the major public health problems worldwide (Mohamed et al., 2018). Several antibiotics have been used to treat the infections caused by these bacteria based on the Clinical & Laboratory Standards Institute: CLSI Guidelines (CLSI) including b-lactams such as carbapenem, aminoclycosides, quinolones, and lipopeptides (Institute–CLSI, 2017).
Carbapenem is a broad-spectrum antibiotic (Mohamed et al., 2018). It is also one of the most potent drugs used to treat infections caused by these MDR-P. aeruginosa (Mirsalehian et al., 2017). However, emergence of carbapenem-resistant (CR) strains have launched a major health issue around the world (Dogonchi et al., 2018).
The antibiotic resistance of P. aeruginosa is either a self-derived process or induced by genetic material transferring using factors such as plasmids. The resistance can be initiate by certain mechanisms such as decreased antibiotic uptake due to reduced outer-cellular-membrane permeability, increased drug pumping through efflux pump overactivity, bacterial change of the antibiotic target site, and the production of carbapenemases, a potent member of the b-lactamase group (Bassetti et al., 2018). Using Ambler classification, carbabenemase enzymes are classified according to their molecular properties to A, B, and D, where A and D require a serine, namely serine carbabenemase (SCMs), in their active sites to perform reactivity, while Class B, called metallo-b-lactamases (MBLs), requires Zn++ to break down the b-lactam ring (Jean et al., 2015; Bonomo, 2017; Khan, Maryam and Zarrilli, 2017).
The present study aimed at investigating the spread of serine carbapenemase-producing isolates of Pseudomonas aeruginosa recovered from some hospitals and specialized health centers in Al-Diwaniyah City, Iraq.
Patients and sample collection
In the current work, 630 clinical samples (244 burn swabs, 163 sputa, 115 urine samples, and 108 ear swabs) were collected from patients during July to December, 2018, from different hospitals and specialized health centers in Al-Diwaniyah province, Iraq. Highly aseptic conditions were followed during the sampling. The samples were then ice-box transported to a microbiological laboratory in the University of Al-Qadisiyah, Diwaniyah City, Iraq, to perform the required tests.
Cultivation, identification, sensitivity tests
MacConky and blood agars were used to grow the bacterium. The morphological and biochemical tests, API 20 E, and Vitek 2 system were utilized to recognize the identity of the bacterium. These tests were followed methods mentioned by (MacFaddin, 2000; Benson, 2002; Dulczak and Kirk, 2005; Winn, 2006; Brown, 2009; Fritsche et al., 2011). The sensitivity method used was Kirby-Bauer and was performed according to (Institute–CLSI, 2017). The antibiotic sensitivity test discs (Bioanlyse Company) used in the present work were ticarcillin-clavulanat (CTC), piperacillin-tazobactam (PTZ), ciprofloxacin (CIP), levofloxacin (LEV), cefepime (FEP), ceftazidime (CAZ), imipenem (IMP), meropenem (MEM), aztreonam (ATM), amikacin (AK), gentamicin (CN), tobramycin (TOB), netilmicin (NET), Colisten (CT), and Polymyxin B (PB).
Molecular detection
Genomic DNA extraction
The DNA was extracted using a specialized kit for genomic DNA extraction manufactured by (Geneaid, USA). The process of the extraction was performed following the kit protocol provided by the mentioned company. The DNA was scanned using a NanoDrop to evaluate the quantity and the quality of the extracted DNA. The resulted DNA was stored in a deep freezer waiting to perform the next tests.
Monoplex polymerase chain reaction
A monoplex polymerase chain reaction (MPCR) technique targeting 4 different serine carbapenemase (SCMs) coding genes was used. The primers used are mentioned in table (1).
Table (1):
The primers of the PCR
Primer |
Sequence (5→3) |
Product size (bp) |
Reference |
|
|---|---|---|---|---|
OXA-48 |
F |
GCG TGG TTA AGG ATG AAC AC |
438 |
(Poirel et al., 2011) |
R |
CAT CAA GTT CAA CCC AAC CG |
|||
GES |
F |
TCA CTC TGC ATA TGC GTC GG |
792 |
(Murugan et al., 2018) |
R |
ACT TGA CCG ACA GAG GCA AC |
|||
KPC |
F |
ATG TCA CTG TAT CGC CGT CT |
893 |
(Bradford et al., 2004) |
R |
TTT TCA GAG CCT TAC TGC CC |
|||
SME |
F |
ACT TTG ATG GGA GGAT TGG C |
551 |
(Poirel et al., 2011) |
R |
ACG AAT TCG AGA TCA CCA G |
The PCR reactional solution was prepared using Accu power PCR Pre Mix (Bioneer Company, Korea). The solution was initiated with the use of 5µl DNA template, 2µl of each primer (F or R), and 11µl PCR water to complete the total volume, 20µl. The thermocycler conditions are provided in table (2).
Table (2):
The thermocycler reaction conditions
| Gene | Time and temperature (°C) | No. of | ||||
|---|---|---|---|---|---|---|
| Initial | Conditions used | Final | cycles | |||
|
|
denaturation | Denaturation | Annealing | Extension | extension | |
| blaKPC | 94/5 min | 94/30s | 57/30s | 72/45s | 72/10min | 30 |
| blaGES | 94/5 min | 94/30s | 59/30s | 72/1.5min | 72/7min | 35 |
| blaOXA-48 | 94/5 min | 94/5min | 57/30s | 72/30min | 72/5min | 35 |
| blaSME | 94/10 min | 94/30s | 60/30s | 72/50s | 72/5min | 36 |
The PCR products were electrophoresed through a 1%-agarose gel pretreated with ethidium bromide. The PCR products were then visualized under a UV imager.
Phylogenetic analysis
The DNA sequencing was performed to determine the nucleotide sequence of the carbapenemase coding genes. The PCR products were sent out to do sequencing by Macrogen Company, South Korea. The sequencing data were analyzed using the NCBI-based BLAST website, and the MEGA v6.0 was used to construct the phylogenetic tree based on the Unweighted Pair Group Methods with Arithmetic Mean (UPGMA tree) method.
Statistical analysis
The data of the study were statistically analyzed using Statistical Package for Social Science (SPSS), 23rd edition. Chi-square test was used to confirm the differences in the data. Confidence interval was equal to 95%, Probability ≤ 0.05 (P≤0.05).
The results of the phenotypic examination showed that the bacterial colonies were pale in color on The MacConky agar as no lactose fermentation was induced, a grape-like odor, and b-hemolysis on the blood agar. Microscopic diagnosis of bacteria showed that the bacteria were motile Gram –ve short rods. The results of the biochemical tests showed that the bacteria were positive for catalase, oxides, citrate, motility, and gelatin liquefaction. The API 20 E and Vitek 2 system showed that 100 of the isolates belonged to P. aeruginosa, Fig. 1.
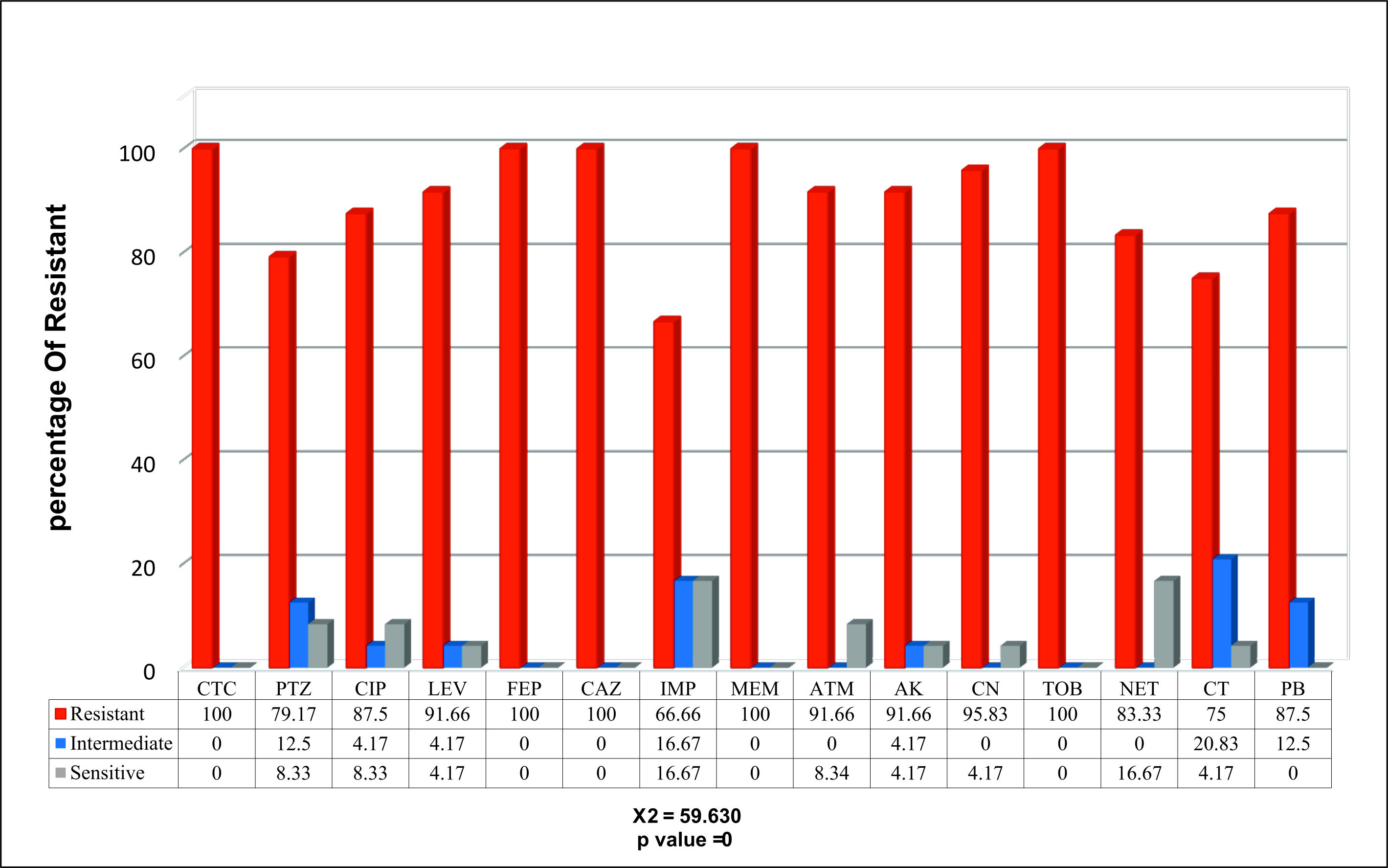
With different severity of resistance by percentages of isolates to various antibiotics, only 24% of the isolates displayed resistance against carbapenem, Fig. 2.
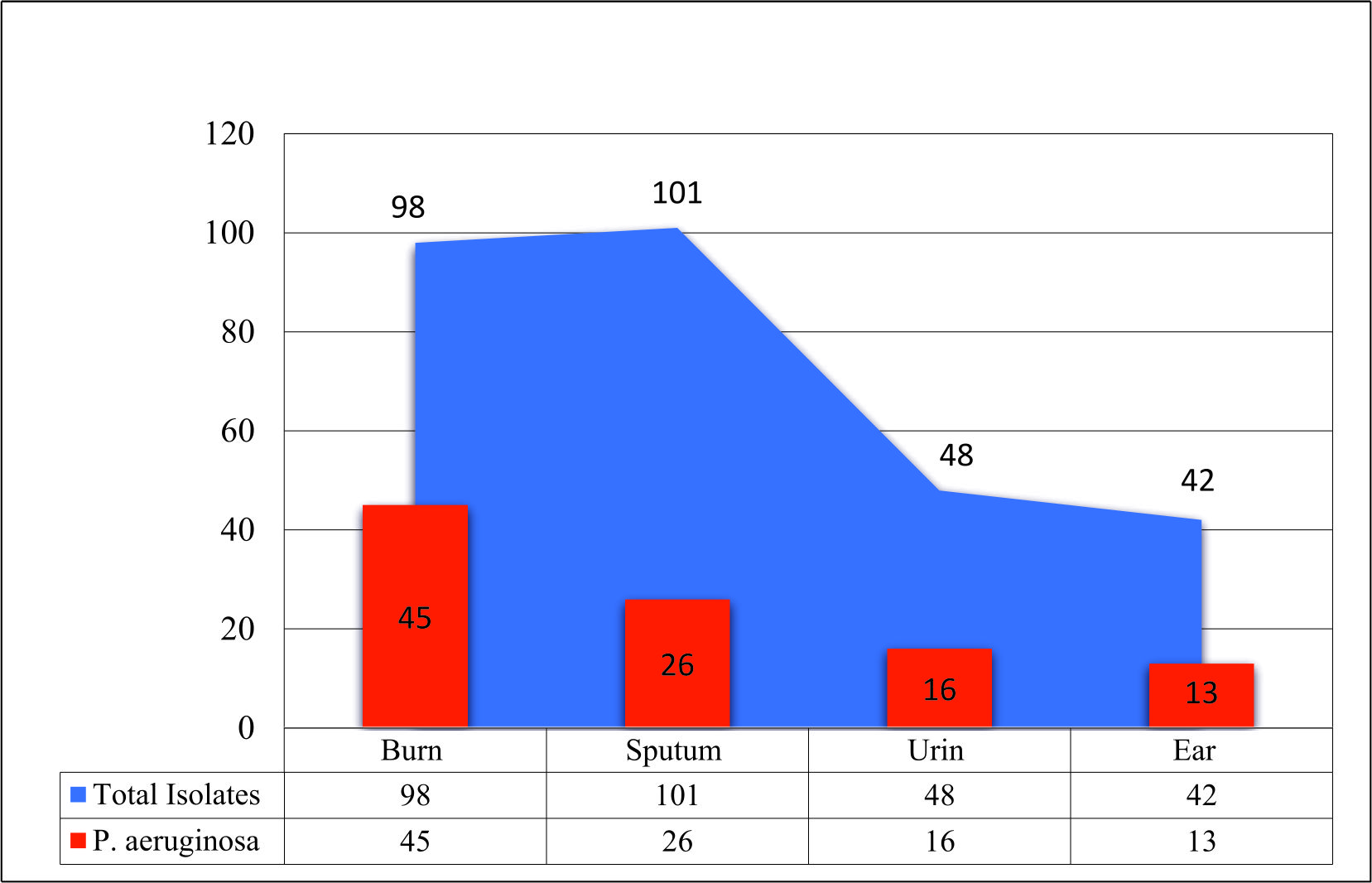 Fig. 2. The percentages of the sensitivity test for the resistant P. aeruginosa isolates to carbapenem based on the CLSI (Institute-CLSI, 2017)
Fig. 2. The percentages of the sensitivity test for the resistant P. aeruginosa isolates to carbapenem based on the CLSI (Institute-CLSI, 2017) The results of the SCM-related MPCR showed the occurrence of the blaGES, blaSME, blaKPC, and blaOXA-48 genes in 20 (83.3%), 19 (79.1%), 11 (45.8%), 18 (75%), respectively, of the isolates, Figs. 3, 4, 5, and 6.
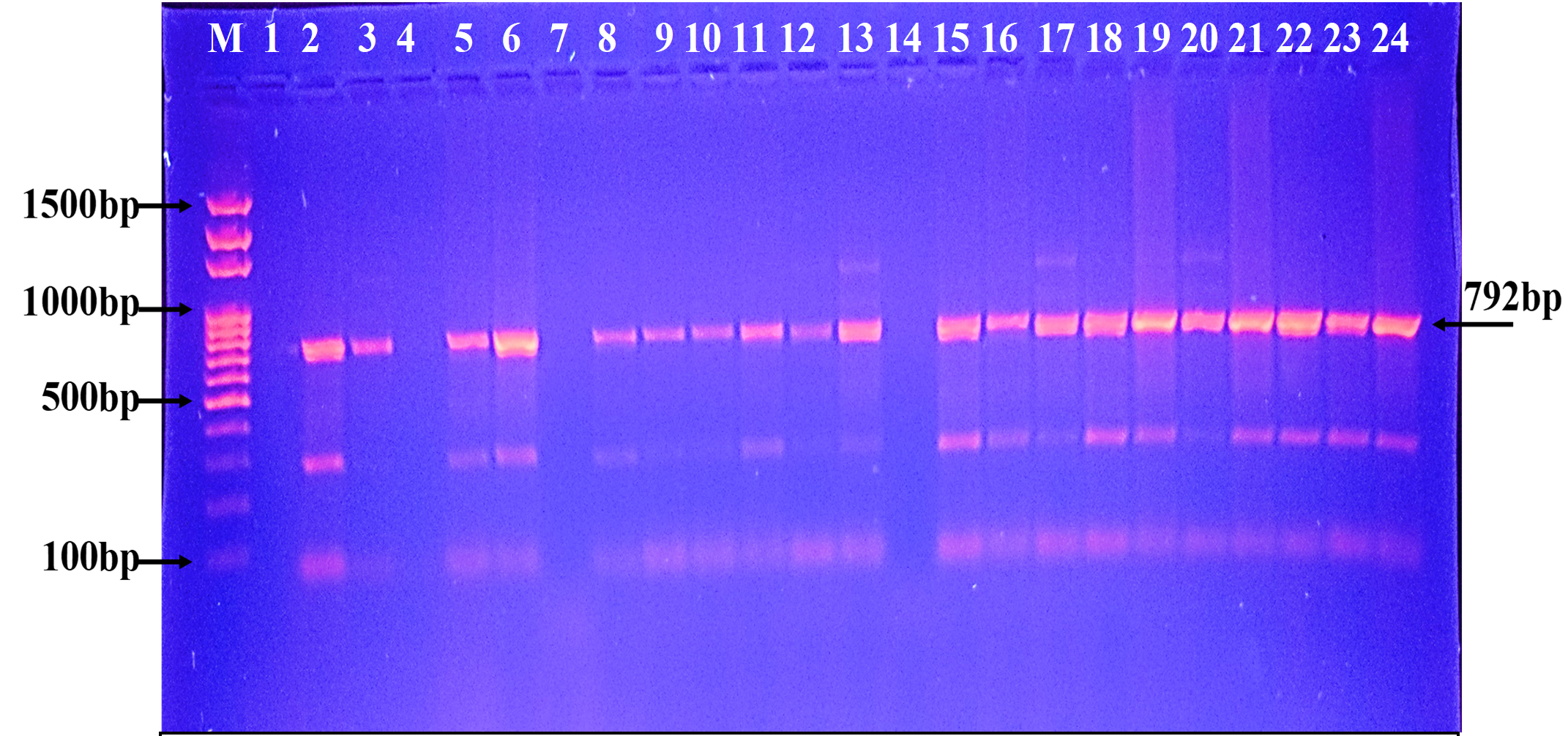 Fig. 3. Agarose gel electrophoresis of the blaGES gene the resistant P. aeruginosa isolates to carbapenem. M: ladder, 1500-100bp. Other lanes: positive amplification, 792bp
Fig. 3. Agarose gel electrophoresis of the blaGES gene the resistant P. aeruginosa isolates to carbapenem. M: ladder, 1500-100bp. Other lanes: positive amplification, 792bp  Fig. 4. Agarose gel electrophoresis of the blaSME gene the resistant P. aeruginosa isolates to carbapenem. M: ladder, 1500-100bp. Other lanes: positive amplification, 551bp
Fig. 4. Agarose gel electrophoresis of the blaSME gene the resistant P. aeruginosa isolates to carbapenem. M: ladder, 1500-100bp. Other lanes: positive amplification, 551bp 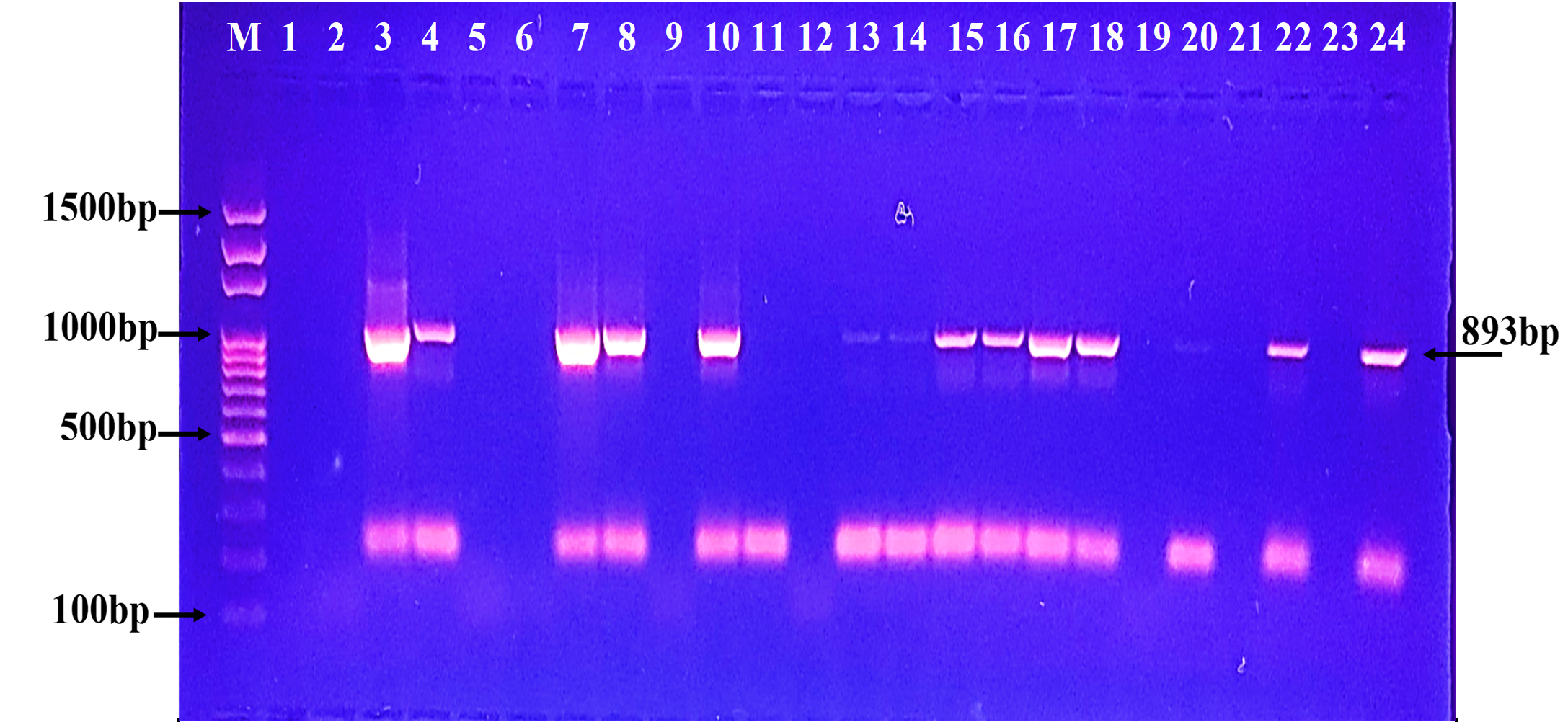 Fig. 5. Agarose gel electrophoresis of the blaKPC gene the resistant P. aeruginosa isolates to carbapenem. M: ladder, 1500-100bp. Other lanes: positive amplification, 893bp
Fig. 5. Agarose gel electrophoresis of the blaKPC gene the resistant P. aeruginosa isolates to carbapenem. M: ladder, 1500-100bp. Other lanes: positive amplification, 893bp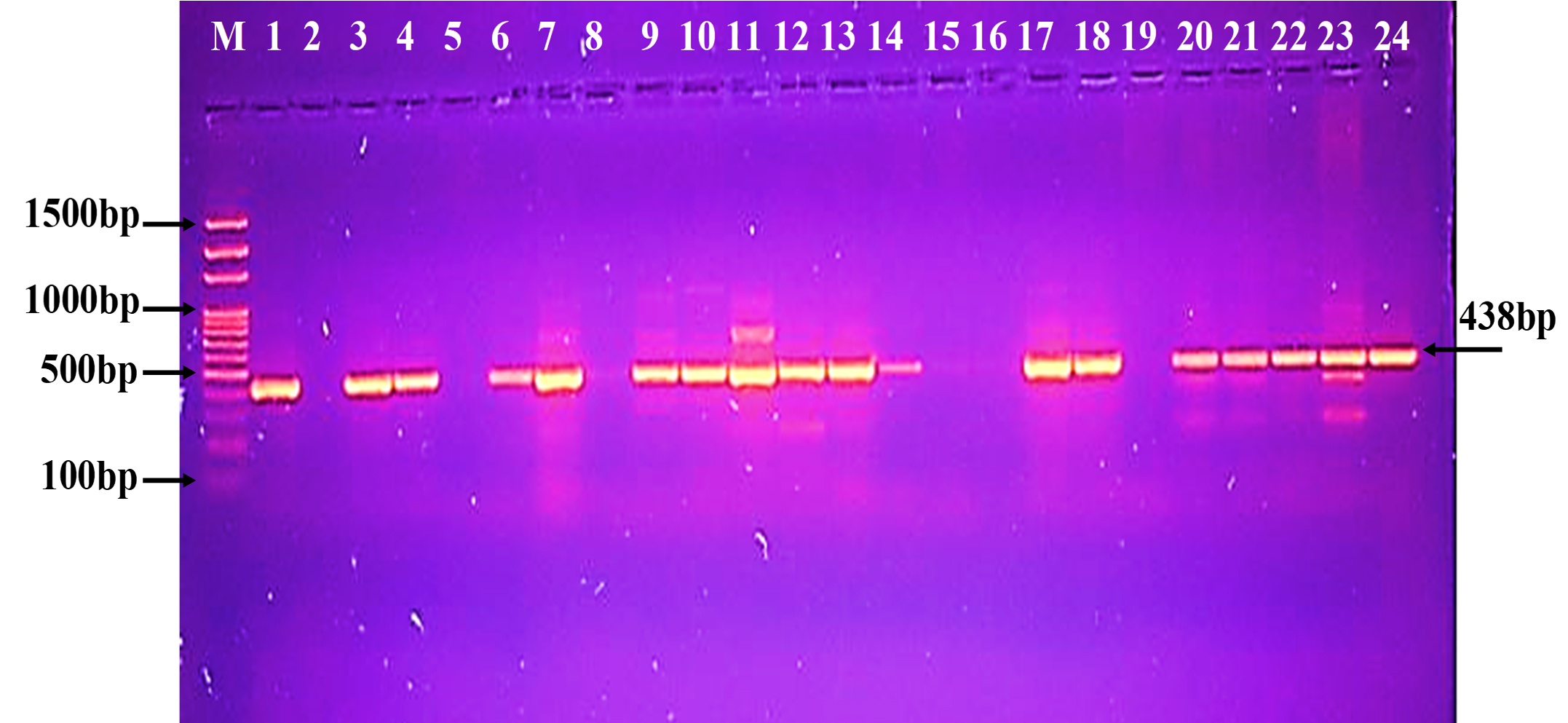 Fig. 6. Agarose gel electrophoresis of the bla (OXA-48) gene the resistant P. aeruginosa isolates to carbapenem. M: ladder, 1500-100bp. Other lanes: positive amplification, 438bp
Fig. 6. Agarose gel electrophoresis of the bla (OXA-48) gene the resistant P. aeruginosa isolates to carbapenem. M: ladder, 1500-100bp. Other lanes: positive amplification, 438bp The phylogenetic study analysis revealed high, 99%, identity with NCBI-related world strains, table 3 and Fig. 7.
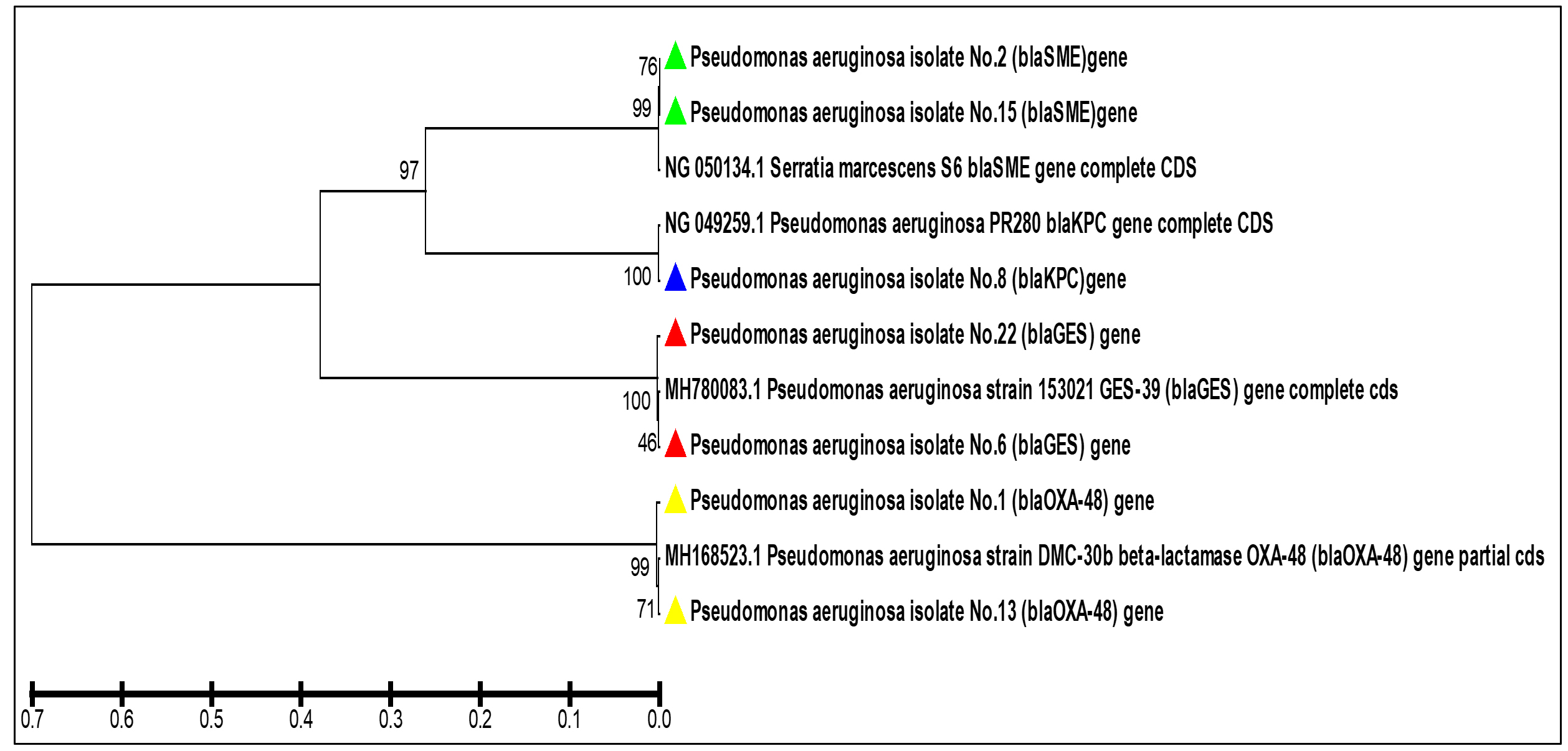 Fig. 7. The phylogenetic tree based on the partial gene sequencing of the serine carbapenemase genes in the resistant P. aeruginosa isolates to carbapenem
Fig. 7. The phylogenetic tree based on the partial gene sequencing of the serine carbapenemase genes in the resistant P. aeruginosa isolates to carbapenemTable (3):
The NCBI-BLAST Homology Sequence identity for the current study isolates of the
P. aeruginosa resistant to carbapenem
| aeruginosa | resistance | Genbank | Isolate | Genbank | (%) | SNP |
|---|---|---|---|---|---|---|
| local isolate | gene | Accession | Accession | |||
| number | number | |||||
|
P. aeruginosa |
blaGES-39 | MN182483 | Pseudomonas aeruginosa | MH780083.1 | 99% | C/A, C/G |
| isolate No.6 | strain 153021 | |||||
| P. aeruginosa | blaGES-39 | MN182482 | Pseudomonas aeruginosa | MH780083.1 | 99% | T/A |
| isolate No.22 | strain 153021 | |||||
| P. aeruginosa | blaSME-1 | MN182490 | Serratia marcescens S6 | NG_050134.1 | 99% | G/T |
| isolate No.2 P. aeruginosa | blaSME-1 | MN182491 | Serratia marcescens S6 | NG_050134.1 | 99% | G/T |
| isolate No.15 P. aeruginosa | blaKPC-5 | MN182497 | Pseudomonas aeruginosa | NG_049259.1 | 99% | T/A , G/C, G/T |
| isolate No.8 | PR280 | |||||
| P. aeruginosa | blaOXA-48 | MN182480 | Pseudomonas aeruginosa | MH168523.1 | 99% | G/A, C/G |
| isolate No.1 | strain DMC-30b | |||||
| P. aeruginosa | blaOXA-48 | MN182481 | Pseudomonas aeruginosa | MH168523.1 | 99% | G/A, A/T |
| isolate No.13 strain DMC-30b | ||||||
The results showed that the highest percentage of P. aeruginosa isolates was from the burn swabs (45.91%). Our results agreed with (de Almeida et al., 2017). who detected that P. aeruginosa bacteria were found to be the predominant bacteria in burns. Skin is considered as one of the main mechanical immune barriers that provide protection support against pathogens; however, any destruction facing this barrier may leave the body prone to these opportunistic microorganisms encouraging for nosocomial infections of burns (Russotto et al., 2015; Sousa et al., 2018). The results of the study showed that the most frequent isolated bacteria from burns were the carbapenem-resistant P. aeruginosa (66.67%) recovered from admitted burnt patients who were receiving treatment in the Burn Hospital, Al-Diwaniyah City. This result came in agreement with (Al-Shara, 2013). who revealed that carbapenem-resistant P. aeruginosa isolates were the dominant bacteria isolated from burns.
KPC enzymes are believed to be as one of the most common Class (A) carbapenemases in the world mainly recognized in Klebsiella pneumoniae and Enterobacteriaceae (Francis et al., 2012). In the P. aeruginosa bacteria, the enzymes were first identified in the United States of America in 2010 showing their gene association with the plasmids, allowing them to move horizontally between bacterial species (Poirel et al., 2011). In conducted studies from Iraq, the coding genes of these enzymes were not identified in P. aeruginosa bacteria (Al-Shara, 2013; Mushtak, Shahad and Amer, 2018). Thus and according to the best of our knowledge, the results of the present study reveal for the first time the identification of the bla (KPC) gene in P. aeruginosa bacteria isolated from Iraq. The SME-coding gene was first found on a chromosome in Serratia marcescens in England (1982) (Queenan and Bush, 2007). The enzyme included three types: SME-1, SME-2, and SME-3, identified in different parts of the United States (Queenan and Bush, 2007; Walther-Rasmussen H dnaרiby, 2007). While these enzymes were not previously determined in our country, our results were contrasted with the results of local and global studies to investigate these enzymes in P. aeruginosa isolates (Al-Shara, 2013; Khorvash et al., 2017).
The gene blaGES was the most prevalent; however, the gene has different types such as GES-2 and GES 6, GES-14 and GES-20 (Naas,
Dortet and I Iorga, 2016; Van Duin and Doi, 2017). The coding genes of these enzymes were detected to be carried on a plasmid, allowing for their transferring between bacterial strains and species (Van Duin and Doi, 2017). GES enzymes were identified in P. aeruginosa and A. bumanni in South Africa, Greece, Japan, France, and Korea (Patel and Bonomo, 2013).
In the current work, carbabenemase Class D was identified in P. aeruginosa isolates via the detection of the blaOXA-48 gene. The OXA-48 activity is not limited to carbapenem but is a multi-task enzyme that acts on oxacillin, cloxacillin, and penicillin (Jayol et al., 2016). The enzyme cannot be inhibited by EDTA salt or b-lactams such as Amoxicillin-clavulanic acid (Drawz and Bonomo, 2010) resulting in limited treatment options provided to patients. The OXA-48 enzyme was discovered in bacterial isolates in Egypt, Sudan, Algeria, Tunisia, and France (Nordmann, Naas and Poirel, 2011; Jayol et al., 2016; Mohamed et al., 2018) To the best of our knowledge, there is no studies explored the presence of such enzymes in P. aeruginosa isolates from Iraq. The present study examined the detection of the coding gene of this enzyme bla (OXA-48) in the clinical isolates of carbapenem-resistance P. aeruginosa. The phylogenetic study analysis revealed high, 99%, identity with NCBI-related world strains. This gives valued information that our and the world strains might have descended from the same ancestors or evolved to have those resistance genes via horizontal gene transfer.
According to the best of our knowledge, this is the first study in Iraq that detects a group of encoding genes of serine carbapenemase in carbapenem-resistant isolates. The current work results may enhance developing better control strategies against P. aeruginosa by targeting these genes.
Acknowledgements
We would like to express our thanks to Department of Biology, College of Science, University of AL-Qadisiyah, Iraq.
Conflict of Interest
The authors declares that there is no conflict of interest.
Authors’ Contributions
All authors have made substantial contribution to the work and approved it for publication.
Funding
None.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
Not declared.
- Al-Shara, J. M. R. Phenotypic and molecular detecting of carbapenem resistant Pseudomonas aeruginosa in Najaf Hospitals. Ph. D. Thesis. Sc, University of Kufa. Iraq 2013.
- Alikhani, M. Y., Parsavash, S., Arabestani, M. R., & Mostafa, S. Prevalence of Antibiotic Resistance and Class 1 Integrons in Clinical and Environmental Isolates of Pseudomonas aeruginosa. Avicenna Journal of Clinical Microbiology and Infection,2017, 4(4).
- Bassetti, M., Vena, A., Croxatto, A., Righi, E., & Guery, B. How to manage Pseudomonas aeruginosa infections. Drugs in Context,2018. https://doi.org/10.7573/dic.212527
- Benson, H. J. Bacterial population counts. Microbiological Applications. Laboratory Manual in General Microbiology. 8th Ed.2002. McGraw Hill, New York, USA, 87.
- Bonomo, R. A. ג-Lactamases: a focus on current challenges. Cold Spring Harbor Perspectives in Medicine,2017, 7(1), a025239.
Crossref - Bradford, P. A., Bratu, S., Urban, C., Visalli, M., Mariano, N., Landman, D., … Quale, J. . Emergence of Carbapenem-Resistant Klebsiella Species Possessing the Class A Carbapenem-Hydrolyzing KPC-2 and Inhibitor-Resistant TEM-30 -Lactamases in New York City. Clinical Infectious Diseases,2004, 39(1), 55–60.
Crossref - Brown, A. E. Benson´ s. Microbiological applications: laboratory manual in general microbiology,2009.
- CLSI . Performance Standards for Antimicrobial Susceptibility Testing: 26th Informational Supplement, Document M100-S26. The Clinical and Laboratory Standards Institute, (January), Wayne, PA,2016.
- de Almeida, K. de C. F., Calomino, M. A., Deutsch, G., de Castilho, S. R., de Paula, G. R., Esper, L. M. R., & Teixeira, L. A. Molecular characterization of multidrug-resistant (MDR) Pseudomonas aeruginosa isolated in a burn center. Burns,2017, 43(1), 137–143.
Crossref - Dogonchi, A. A., Ghaemi, E. A., Ardebili, A., Yazdansetad, S., & Pournajaf, A. Metallo-ג-lactamase-mediated resistance among clinical carbapenem-resistant Pseudomonas aeruginosa isolates in northern Iran: A potential threat to clinical therapeutics. Tzu-Chi Medical Journal,2018, 30(2), 90.
Crossref - Drawz, S. M., & Bonomo, R. A. Three decades of ג-lactamase inhibitors. Clinical Microbiology Reviews,2010, 23(1), 160–201.
Crossref - Dulczak, S., & Kirk, J. Overview of the evaluation, diagnosis, and management of urinary tract infections in infants and children. Urol Nurs,2005, 25(3), 185–192.
- Francis, R. O., Wu, F., Della-Latta, P., Shi, J., & Whittier, S. Rapid detection of Klebsiella pneumoniae carbapenemase genes in Enterobacteriaceae directly from blood culture bottles by real-time PCR. American Journal of Clinical Pathology,2012, 137(4), 627–632.
Crossref - Fritsche, T. R., Swoboda, S. E., Olson, B. J., Moore, F. M., Meece, J. K., & Novicki, T. J. . Evaluation of The Sensititre ARIS2x and Vitek 2 Automated Systems for Identification of Bacterial Pathogens Recovered from Veterinary Specimens. Marshfield labs. Lacrosse Univ. Wisconsin. USA, 2011.
- Institute–CLSI, C. & L. S. Performance Standards of Antimicrobial Susceptibility Testing; 27th Informational Supplement; M100-S27. Clinical and Laboratory Standards Institute Wayne, PA, 2017.
- Jayol, A., Poirel, L., Dortet, L., & Nordmann, P. National survey of colistin resistance among carbapenemase-producing Enterobacteriaceae and outbreak caused by colistin-resistant OXA-48-producing Klebsiella pneumoniae, France, 2014. Eurosurveill,2016, 21(37).
Crossref - Jean, S.-S., Lee, W.-S., Lam, C., Hsu, C.-W., Chen, R.-J., & Hsueh, P.-R. Carbapenemase-producing Gram-negative bacteria: current epidemics, antimicrobial susceptibility and treatment options. Future Microbiology,2015, 10(3), 407–425.
Crossref - Khan, A. U., Maryam, L., & Zarrilli, R. Structure, genetics and worldwide spread of New Delhi metallo-ג-lactamase (NDM): a threat to public health. BMC Microbiology, 2017, 17(1), 101.
Crossref - Khorvash, F., Yazdani, M., Shabani, S., & Soudi, A. Pseudomonas aeruginosa-producing Metallo-b-lactamases (VIM, IMP, SME, and AIM) in the Clinical Isolates of Intensive Care Units, a University Hospital in Isfahan, Iran. Advanced Biomed Res.,2017, 6(1), 147.
Crossref - MacFaddin, J. F. Urease test. Biochemical Tests for Identification of Medical Bacteria,2000, 424–438.
- Maroui, I., Barguigua, A., Aboulkacem, A., Elhafa, H., OUARRAK, K., Sbiti, M., Belhaj, A. Clonal Analysis of Clinical and Environmental Pseudomonas aeruginosa Isolates from Meknes Region, Morocco. In Polish Journal of Microbiology ,2017,Vol. 66.
Crossref - Mirsalehian, A., Kalantar-Neyestanaki, D., Taherikalani, M., Jabalameli, F., & Emaneini, M. Determination of carbapenem resistance mechanism in clinical isolates of Pseudomonas aeruginosa isolated from burn patients, in Tehran, Iran. Journal of Epidemiology and Global Health,2017, 7(3), 155–159.
Crossref - Mohamed, S., Alobied, A., Hussien, W., & Saeed, M. blaOXA-48 Carbapenem Resistant Pseudomonas aeruginosa Clinical Isolates in Sudan. Journal of Advances in Microbiology,2018, 10(4), 1–5.
Crossref - Murugan, N., Malathi, J., Therese, K. L., & Madhavan, H. N. Application of six multiplex PCR’s among 200 clinical isolates of Pseudomonas aeruginosa for the detection of 20 drug resistance encoding genes. The Kaohsiung Journal of Medical Sciences,2018, 34(2), 79–88.
Crossref - Mushtak, A. T. S., Shahad, A. A., & Amer, A. Molecular Detection of Medically Important. Asian Journal of Pharmaceutics,2018, 12(3), 991–1001.
- Naas, T., Dortet, L., & I Iorga, B. Structural and functional aspects of class A carbapenemases. Current Drug Targets,2016, 17(9), 1006–1028.
Crossref - Nordmann, P., Naas, T., & Poirel, L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerging Infectious Diseases,2011, 17(10), 1791.
Crossref - Patel, G., & Bonomo, R. “Stormy waters ahead”: global emergence of carbapenemases. Frontiers in Microbiology,2013, 4, 48.
Crossref - Poirel, L., Walsh, T. R., Cuvillier, V., & Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagnostic Microbiology and Infectious Disease,2011, 70(1), 119–123.
Crossref - Queenan, A. M., & Bush, K. Carbapenemases: the versatile b-lactamases. Clinical Microbiology Reviews,2007, 20(3), 440–458.
Crossref - Russotto, V., Cortegiani, A., Raineri, S. M., & Giarratano, A. Bacterial contamination of inanimate surfaces and equipment in the intensive care unit. Journal of Intensive Care,2015, 3(1), 54.
Crossref - Schaber, J. A., Hammond, A., Carty, N. L., Williams, S. C., Colmer-Hamood, J. A., Burrowes, B. H., … Hamood, A. N. Diversity of biofilms produced by quorum-sensing-deficient clinical isolates of Pseudomonas aeruginosa. Journal of Medical Microbiology,2007, 56(6), 738–748.
Crossref - Sousa, D., Ceniceros, A., Galeiras, R., Pיrtega-Dםaz, S., Gutierrez-Urban, J.-M., Rodriguez-Mayo, M., Llinares, P. Microbiology in burns patients with blood stream infections: trends over time and during the course of hospitalization. Infect. Diseas,2018, 50(4), 289–296.
Crossref - Van Duin, D., & Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence, 2017, 8(4), 460–469.
Crossref - Walther-Rasmussen, J., & Hרiby, N. Class A carbapenemases. J Antimicrob Chemother, 2007, 60(3), 470–482.
Crossref - Winn, WC. Koneman’s color atlas and textbook of diagnostic microbiology. Lippincott williams & wilkins, 2006.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.