ISSN: 0973-7510
E-ISSN: 2581-690X
Johne’s disease (JD), also referred to as paratuberculosis, is caused by Mycobacterium avium subsp. paratuberculosis (MAP). It is a chronic disease of ruminants that primarily affects the enteric system and is seen worldwide. The study aimed towards molecular detection of MAP in bovine samples. A total of 220 bovine samples (intestinal tissue-60, faecal-60 and milk-100) were collected from cattle herds and slaughter point and were examined for detection of MAP targeting the IS900 insertion sequence through conventional PCR. MAP DNA was detected in 7 intestinal tissue samples (11.66%) and 1 faecal sample (1.66%). Only 3/60 (5.0%) intestinal tissue samples could be detected as acid fast bacilli by Z-N staining. The results were further re-confirmed by nBLAST analysis following Sanger dideoxynucleotide sequencing. The results indicate the presence of MAP-infections in dairy herds of the study area, Assam. This may be the first report of molecular detection of MAP in bovine intestinal tissue and faecal samples in Assam, Northeast India as per our literature search. Additional research is required across all Northeastern regions of India to determine the precise prevalence of the pathogen and to mitigate the spread of the disease.
Paratuberculosis, Prevalence, Johne’s Disease, IS900, Acid-fast Bacilli, Sequencing
Johne’s disease (JD) or Paratuberculosis, having a global presence, is a common disease of ruminants with Mycobacterium avium subsp. paratuberculosis (MAP) as the causative agent. The disease is categorized under the List B transmissible diseases by World Organization for Animal Health (WOAH).1 The disease is transmitted mainly by ingestion of the pathogen, i.e. MAP from the contaminated feed, pasture,2 infected colostrum to calves,3 etc. MAP can survive in pasteurization and human can get the infection through contaminated raw milk, meat, and even through contact with infected animals and is debatable for the cause of Crohn’s disease of human.4 There is huge implication in term of production losses also with decreased milk production, wasting, weight loss, and trade restrictions.5
Globally, the prevalence of JD in animal population had been reported from several countries, viz. 15.1% ± 7.3% in sheep from Italy,6 3.7% from Portugal,7 herd level prevalence of 61.3% from Saudi Arabia8 and more recently 0.72% in Brazil,9 etc. JD is endemic in India in ruminant population with its first report from Hisar, Haryana in 1913 but there is lack of proper national surveillance and data.10 Based on tissue smear examination 1% JD prevalence of slaughterhouse specimens was reported from West Bengal.11 There are few other reports on the JD prevalence 6.8% in cattle from Gujarat,12 55% in Punjab13 and 11.7% in Tamil Nadu.14 The JD prevalence in small ruminants had also documented as 24.7% from Madhya Pradesh,15 8.0% in Uttar Pradesh,16 and 29.0% in Rajasthan.10 Infected animals are typically diagnosed with MAP by bacterial isolation and antibody-based assays like ELISA with their own limitation of long incubation period for culture up to 16 weeks and low sensitivity of ELISA during initial phase of infection.17,18
Polymerase chain reaction (PCR) based molecular assay targeting IS900 region is one of the most used assays enabling detection of MAP in early and chronic stages of infection. Hence, the present study was aimed at the detection of insertion sequence IS900 of MAP in bovine samples (milk, faecal and intestinal tissue) by conventional PCR to find out the prevalence of paratuberculosis.
Study design and sample collection
The study was designed in the dairy farms and cattle slaughter points for molecular detection of paratuberculosis involving a total of 220 samples with breakup of milk (n = 100), intestine (n = 60) and fecal (n = 60). Milk samples were collected from dairy cattle, whereas, intestine & fecal samples were collected from slaughtered cattle within the study area. After collection all the samples were brought to laboratory in refrigerated condition and stored in the deep freezer (-20 °C) until its processing. The longitude and latitude of the farms and slaughter points were recorded using (GPS). The present investigation was permitted by Animal Ethics Committee, Assam Agriculture University (770/GO/Re/S/03/CPCSEA/FVSc/AAU/IAEC/19-20/739 dt. 23.12.2019).
Sample processing
Milk samples were processed for molecular detection by following the method of Dundee et al.19 and the tissue materials were processed by following the protocol of van Ingen et al.20 The tissue samples were primarily screened for acid-fast bacteria by Ziehl-Neelsen staining, and were subjected to molecular detection.
Genomic DNA extraction
DNA was extracted from the milk and tissue samples via the Qiagen DNeasy® Blood & Tissue Kit (Cat. No./ID: 69504) by following the manufacturer’s instructions. DNA extraction from the faecal samples were carried out using power fecal DNA extraction kit (QIAamp® PowerFecal® DNA Kit) as per the manufacturer’s instructions. Extracted samples were stored at -20 °C for further molecular analysis.
Polymerase chain reaction (PCR) for detection of MAP
The extracted DNA from the samples (milk, fecal and intestine) were subjected for molecular detection of MAP using IS900 gene based PCR.21 The forward primer sequence was 5′-CCGCTAATTGAGAGATGCGATTGG-3′ and reverse primer sequence was 5′-AATCAACTCCAGCAGCAGCGCGGCCTCG-3′ amplifying 229 bp target of IS900 with conditions viz., initial denaturation: 95 °C for 5 min, 40 repetitions (denaturation: 95°C/35sec , annealing: 62 °C/30 sec, extension: 72 °C/35 sec) and final extension of 72 °C/7 min). We have standardized the IS900 PCR using positive MAP DNA supplied by our research collaborator and used it as positive control for further reactions. All reactions were carried out in a thin-walled 200 μL PCR tube. Ready-to-use solution of DreamTaq Green PCR Master Mix (2X) (Thermo Scientific, Massachusetts, United States) was used in the study containing DreamTaq DNA Polymerase, dNTPs, MgCl2 and optimized buffer. The total reaction volume was made for 20 μL with 10 μL Master Mix, 2.0 μL of template DNA, 1.0 μL each primer (10 pmoL), and nuclease free water (6 μL). The standardization was done for the primers with positive and negative DNA controls and then subsequently used for the samples. The positive samples for IS900 were re-confirmed through Sanger’s sequencing. The sequenced product was subjected to BLAST analysis (NCBI BLASTn).
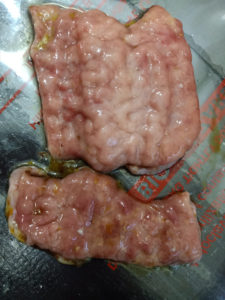
In this study, a variety of diagnostic procedures was performed to determine the presence of MAP in suspected animals, including smears from faecal and intestinal lesions, necropsy, and molecular detection in faeces, milk, and tissues. Gross pathological examination of the intestinal tissue samples revealed enlarged mesenteric and ileocolic lymph nodes with mild to moderate diffuse thickening of the intestinal mucosa. Brain-like corrugation with granular mucosa have also been seen in the small intestine of some dairy cows with significantly thickened intestinal wall and mucosal folds (Figure 1). The miliary granulomatous lesions occasionally protruded from the small intestinal wall in seriously distressed animals. Inside the intestinal wall, pus-like granulomatous lesions were observed, along with delicate mucosal folds.
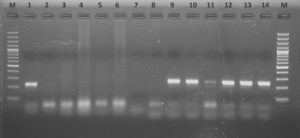
In the present study, out of 60 intestinal tissue samples analyzed, 3/60 (5.0%) samples were detected as acid fast bacilli under microscopic examination and 7/60(11.66%) could amplify 229 bp of MAP DNA by IS900 PCR (Figure 2) and their identifications were confirmed by sequencing. Out of 60 faecal samples, only 1/60 (1.66%) sample showed positivity for MAP DNA by IS900 PCR. Out of 100 milk samples analyzed, no samples could be amplified for MAP DNA in PCR.
Figure 2. Amplified 229 bp of the IS900 gene product of positive tissue samples in 1.8% agarose gel L-Ladder (Thermo Scientific GeneRuler 100 bp plus DNA Ladder); Lane 1-positive control; Lane 2-negative control Lane 3-14: Samples
Sequence based confirmation of the sample was undertaken by ascertaining the sequence of the amplicons by Sanger dideoxy sequencing. The obtained sequences were checked for quality and blasted into NCBI-BLAST database. The first hit was taken as the identification result. The positive sample of IS900 gene revealed 94% similarity with species deposited in GenBank, identifying Mycobacterium avium subsp. paratuberculosis. The sequence was submitted to NCBI and accession number was obtained (OQ417943.1). Further phylogenetic analysis was undertaken by a multiple sequence alignment method in MEGA X software. Aligned sequences were chosen from various geographical locations for a spatial comparison. The sequences were aligned using MUSCLE algorithm and phylogeny was constructed using Neighbour-End Joining (NEJ) method with 1000 bootstrap replicates. Maximum Composite Likelihood method was used to compute the evolutionary distances. IS900 were used successfully for sequencing in bovine clinical sample for assigning its unique stand in the dendrogram. The dendrogram is presented, which were constructed to represent the species identification through BLAST in a defined manner by placing the sample under a single tree along with their respective global and standard isolates (Figure 3).
The positivity of the targeted animals and samples as read out with post-mortem observation, smear microscopy and IS900 PCR could prove that Johne’s disease does exist in the cattle population in the study area. The occurrence of 11.66% positivity in IS900 based PCR shows that there is a significant number of MAP infected animals within and around the Guwahati city. The affected animals were highly emaciated, with complete loss of adipose tissue. On postmortem examination, the mesenteric and ileo-colic lymph nodes in the small intestine were moderately enlarged with brain-like corrugation have been seen in the small intestine of some dairy cows with substantially thickened intestinal wall. Other scientists also recorded similar type of observations, describing the small intestine as having a substantially thickening of intestinal wall, corrugation and granular mucosa.22-24 Perez-Rodriguez et al.25 also reported pathological features in caprines with granulomatous inflammation in the small intestines and mesenteric lymph nodes.
In this study, MAP could not be detected in the milk, which might be due to the low prevalence or lower sensitivity of PCR for milk samples or due to intermittent release of Mycobacteria in milk during a short period post-infection, etc.26 Fecal samples had a lower number of positive cases in molecular detection than intestinal samples in our investigation. The intestinal samples have visible lesions which contain more numbers of pathogen than fecal shedding is one of the reasons and the faecal samples contain inhibitory substances that can impede DNA purity affecting the downstream PCR amplification. Still fecal PCR is one of the better options in live animal than culture which take very long incubation time.27,28 Verma et al.29 also detected MAP from fecal samples of cattle from Jabalpur, Madhya Pradesh with IS900 based PCR assay. It was found in our study that IS900 based molecular assay has given a good and accurate results regarding the diagnosis of paratuberculosis from direct sample. In live animals, fecal-based PCR detection is a rapid, non-invasive method that requires minimal handling and can provide a preliminary indication of the presence of the disease within the herd.30 For individual animal screening, the rectal pinch method is considered more reliable for sample collection. In post-mortem cases, intestinal samples showing visible signs such as corrugation or miliary nodules are ideal for testing. These samples not only offer a fair indication of disease presence but can also assist in tracing the origin of the infected animal or the source farm/location. Besides these molecular diagnostic assays, ELISA-based serological assays are also available for diagnosis of Johne’s diseases in bovines.31 There are few publications on paratuberculosis in India due to a lack of ongoing surveillance and testing facilities. The goal of this study was to compare ZN staining and molecular assays to evaluate a diagnostic method for MAP in a specific study region. Molecular assays can give valuable observations and inferences at the individual animal and herd levels.
The findings of current investigation provide evidence about the presence of paratuberculosis infection in the cattle population of Guwahati Metropolitan City of Assam, India. The IS900 PCR based direct detection of MAP from tissue and faecal materials could serve as a valuable diagnostic or screening test for herds with JD. The present finding was the first attempt to diagnosis the disease by molecular assay in the study area and an attempt for a systematic epidemiological study of paratuberculosis in Assam, India. Further detailed research and investigations in this direction is warranted to assess the exact scale of the burden in the state and region, so as to formulate future strategies for limiting the spread of this dreaded disease in both animals and humans.
ACKNOWLEDGMENTS
The authors sincerely thank the Dean, College of Veterinary Science, Assam Agricultural University, and the Director, ICAR Research Complex for NEH Region, Umiam, for their support and permission to conduct this collaborative study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study is supported by the College of Veterinary Sciences, Assam Agriculture University, India, and ICAR RC NEH, Umiam, Meghalaya, India.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Animal Ethics Committee, College of Veterinary Science, AAU, Khanapara, vide approval number-770/GO/Re/S/03/CPCSEA/FVSc/AAU/ IAEC/19-20/739 dated 23.12.2019.
- OIE. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; Paratuberculosis (Johne’s disease) – Chapter 3.1.17. World Organisation for Animal Health, Paris, France. 2024. https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.01.17_PARATB.pdf
- Lavers CJ, McKenna SLB, Dohoo IR, Barkema HW, Keefe GP. Evaluation of environmental fecal culture for Mycobacterium avium subsp. paratuberculosis detection in dairy herds and association with apparent within-herd prevalence. Can Vet J. 2013;54(11):1053-1060.
- Fecteau ME, Whitlock RH, Buergelt CD, Sweeney RW. Exposure of young dairy cattle to Mycobacterium avium subsp. paratuberculosis (MAP) through intensive grazing of contaminated pastures in a herd positive for Johne’s disease. Can Vet J. 2010;51(2):198-200.
- Eltholth MM, Marsh VR, Van Winden S, Guitian FJ. Contamination of food products with Mycobacterium avium paratuberculosis: a systematic review. J Appl Microbiol. 2009;107(4):1061-1071.
Crossref - Vazquez P, Garrido JM, Juste RA. Effects of paratuberculosis on Friesian cattle carcass weight and age at culling. Span J Agric Res. 2012;10(3):662-670.
Crossref - Attilli RA, Victor NN, Silvia P, Luciana P, Anastasia D, Vincenzo C. Ovine paratuberculosis: a seroprevalence study in dairy flocks reared in the Marche region, Italy. Vet Med Int 2011;2011:782875.
Crossref - Coelho AC, Pinto ML, Coelho AM, Aires A, Rodrigues J. A Seroepidemiological survey of Mycobacterium avium subsp. paratuberculosis in sheep from North of Portugal. Pesq Vet Bras. 2010;30(11):903-908.
Crossref - Elsohaby I, Fayez M, Alkafafy M, et al. Serological and molecular characterization of Mycobacterium avium subsp. paratuberculosis (MAP) from sheep, goats, cattle and camels in the Eastern Province, Saudi Arabia. Animals. 2021;11(2):1-11.
Crossref - de Noronha Xavier A, de Sa Luenda MN, de Nazare SFM, et al. First serological diagnosis of Mycobacterium avium subsp. paratuberculosis infection in sheep in the state of Pernambuco, Brazil. Vet Res Commun. 2024;48(2):1293-1299.
Crossref - Singh SV, Singh AV, Singh R et al. Sero-prevalence of Bovine Johne’s disease in buffaloes and cattle population of North India using indigenous ELISA kit based on native Mycobacterium avium sub species paratuberculosis ‘Bison type’genotype of goat origin. Comp Immunol Microbiol Infect Dis. 2008;31(5):419-433.
Crossref - Mukerji A, Lahiri A. Investigation of Johne’s disease in buffaloes. Indian Vet J. 1960;37:349-353.
- Mohan MS, Duraisamy P, Praveena PE, et al. Prevalence of paratuberculosis in cattle and buffaloes. Indian Vet J. 2009;86(1):4-6.
- Kaur P, Filia G, Singh SV, Patil PK, Kumar GVPPSR, Sandhu KS. Molecular epidemiology of Mycobacterium avium subspecies paratuberculosis: IS900 PCR identification and IS1311 polymorphism analysis from ruminants in the Punjab region of India. Comp Immunol Microbiol Infect Dis. 2011;34(2):163-169.
Crossref - VinodhKumar OR, Gunaseelan L, Ronald BSM, Sakthivelan SM. Slaughterhouse prevalence of ovine paratuberculosis in Southern India. Trop Anim Health Prod. 2013;45(4):1063-1069.
Crossref - Rajukumar K, Tripathi BN, Kurade NP, Parihar NS. An enzyme-linked immunosorbent assay using immono affinity-purified antigen in the diagnosis of caprine paratuberculosis and its comparison with conventional ELISAs. Vet Res Commun. 2001;25(7):539-553.
Crossref - Singh SV, Singh AV, Singh PK, Kumar A, Singh B. Molecular identification and characterization of Mycobacterium avium sub species paratuberculosis in free living non-human primate (Rhesus macaques) from North India. Comp Immunol Microbiol Infect Dis. 2011;34(3):267-271.
Crossref - Collins DM, Radford AJ, de Lisle GW, Billiman-Jacobe H. Diagnosis and epidemiology of bovine tuberculosis using molecular biological approaches. Vet Microbiol. 1994;40(1-2):83-94.
Crossref - O’mahony J, Hill C. Rapid real-time PCR assay for detection and quantitation of Mycobacterium avium subsp. paratuberculosis DNA in artificially contaminated milk. Appl Environ Microbiol. 2004;70(8):4561-4568.
Crossref - Dundee L, Grant IR, Ball HJ, Rowe MT. Comparative evaluation of fourdecontamination protocols for the isolation of Mycobacterium avium subsp. paratuberculosis from milk. Lett Appl Microbiol. 2001;33(3):173-177.
Crossref - van Ingen J, Blaak H, de Beer J, de Roda Husman AM, van Soolingen D. Rapidly growing nontuberculous mycobacteria cultured from home tap and shower water. Appl Environ Microbiol. 2010;76(17):6017-6019.
Crossref - Vary PH, Andersen PR, Green E, Hermon-Taylor J, McFadden JJ. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne’s disease. J Clin Microbiol.1990;28(5):933-937.
Crossref - Mobius P, Liebler-Tenorio E, Holzer M, Kohler H. Evaluation of associations between genotypes of Mycobacterium avium subsp. paratuberculsis and presence of intestinal lesions characteristic of paratuberculosis. Vet Microbiol. 2017;201:188-194.
Crossref - Field N L, McAloon C G, Gavey L, Mee JF. Mycobacterium avium subspecies paratuberculosis infection in cattle – a review in the context of seasonal pasture-based dairy herds. Ir Vet J. 2022;75(1):12.
Crossref - Mohammed T, Mamo G, Zewude A, Sirak A, Gumi B, Ameni G. Prevalence of paratuberculosis in cattle based on gross and microscopic lesions in Ethiopia. BMC Vet Res. 2023;19(1):203.
Crossref - Perez-Rodriguez E, Walton IPJ, Hernandez JJS, et al. ADA1ADAp ratio in pleural tuberculosis: an excellent diagnostic parameter in pleural fluid. Respir Med. 1999;93(11):816-821.
Crossref - Carvalho RCT, Castro VS, Fernandes DVGS, et al. Use of PCR for detection of bovine tuberculosis bacillus in milk of positive skin test cows. Braz J Vet Res Anim Sci. 2014;51(1):42-48.
Crossref - Merkal RS, Whipple DL, Sacks JM, Snyder GR. Prevalence of Mycobacterium paratuberculosis in ileocecal lymph nodes of cattle culled in the United States. J Am Vet Med Assoc. 1987;190(6):676-680.
Crossref - Stabel JR, Wells SJ, Wagner BA. Relationships between fecal culture, ELISA, and bulk tank milk test results for Johne’s disease in US dairy herds. J Dairy Sci. 2002;85(3):525-531.
Crossref - Verma YK, Verma Y, Swamy M, et al. Molecular and serological detection of Mycobacterium avium subspecies paratuberculosis in ruminants of Jabalpur region. Ind J Ani Res. 2024:B-5149.
Crossref - Wichert A, Einax E, Hahn N, et al. Detection of Mycobacterium avium Sub species Paratuberculosis in Pooled Fecal Samples by Fecal Culture and Real-Time PCR in Relation to Bacterial Density. Animals. 2021;11(6):1605.
Crossref - Elsohaby I, Kostoulas P, Fayez M, et al. Bayesian estimation of diagnostic accuracy of fecal smears, fecal PCR and serum ELISA for detecting Mycobacterium avium subsp. paratuberculosis infections in four domestic ruminant species in Saudi Arabia. Vet Microbiol. 2025;301:110377.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.