ISSN: 0973-7510
E-ISSN: 2581-690X
Pseudomonas aeruginosa producing extended spectrum beta lactamases (ESBL) is a major concern in the hospital settings. It is usually reported in Enterobacteriaceae and is less frequently observed in P. aeruginosa. There is no recommended test for ESBL detection in P.aeruginosa. Therefore, we determined the occurrence of ESBL in clinical isolates of P.aeruginosa by both phenotypic and genotypic methods. Antimicrobial susceptibility tests were done on two hundred and thirteen isolates of P. aeruginosa. Phenotypic detection of ESBL was performed using combined disk method and ESBL encoding genes such as blaVEB, blaPER, blaPSE, blaGES, blaTEM, blaSHV, blaCTX-M, blaBEL, blaOXA1, blaOXA10, blaOXA2 were studied by simplex PCR. Of the 213 isolates, 85 were identified as resistant to ceftazidime and 27/85 isolates were confirmed to be ESBL producers by phenotypic method. The presence of genes encoding ESBLs comprising of blaTEM (n=44), blaOXA-10 (n=19) isolates, blaOXA-1 (n=5), blaOXA-2 (n=3) were found. All OXA gene positive isolates exhibited the ESBL phenotype. The blaGES gene were identified in 4/85 (5%) isolates. This study shows the prevalence of ESBL among clinical isolates of P.aeruginosa and in particular, the presence of GES β lactamases.
Extended Spectrum β-Lactamases, Class D β-Lactamases, Pseudomonas aeruginosa, PCR
Pseudomonas aeruginosa is a multidrug resistant bacterium which has intrinsic resistant mechanisms and is responsible for hospital acquired infections. The production of beta-lactamases is the most common mechanism of bacterial antimicrobial resistance, which is primarily mediated by plasmids.1 Extended spectrum beta lactamases (ESBL) are a group of β lactamases which hydrolyze penicillins and cephalosporins including oxyimino b-lactamases and aztreonam, but are inhibited by β-lactamase inhibitors. Co-resistance to many additional antibiotic classes is common in organisms that produce ESBLs, thereby limiting therapeutic options. Penicillinases belonging to the molecular class A serine β lactamases (CTX-M families, TEM and SHV) and class D (OXA-type) β lactamases are the commonly reported β-lactamases; VEB, PSE, CARB, PER and GES are less frequently reported.2 In addition, Insertion sequence (IS) elements are closely linked to blaESBL genes. Insertion Sequence 26 (IS26) is located upstream of blaTEM gene and is frequently reported to reside in a resistance plasmid which has the feature of transposition and target site duplication (TSD).3 IS26 is significant in the acquisition and dissemination of antibiotic resistance gene. The overexpression of inducible chromosomal AmpC β-lactamases in P. aeruginosa confers resistance to broad spectrum antibiotics which may be difficult to detect by phenotypic methods.4 Increased membrane permeability and the existence of several efflux systems adds to the difficulties in detection of antibiotic resistance. There is no reliable test for phenotypic detection of ESBLs in clinical isolates of P. aeruginosa. Molecular methods are used to detect antibiotic resistance genes for monitoring the emergence of drug resistance in the clinical setting. The detection of these genes could help to establish standards for hospital infection control measures. Hence the study was aimed to detect ESBL production by both phenotypic and genotypic methods.
During the period of one year (2013-2014), 213 Pseudomonas aeruginosa clinical isolates were collected from two tertiary care hospitals in South India. The ethical clearance was obtained from the institutional human ethics committee (IHEC No: UM/IHEC/02-2014-I). Kirby Bauer disc diffusion method was used to evaluate all clinical isolates for antibiotic susceptibility.5 The following antibiotics (HiMedia Labs Mumbai, India) were used: piperacillin (100 μg), piperacillin/tazobactum (100μg/10 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), ofloxacin (5 µg), aztreonam (30 μg), cefepime (30 μg), ceftazidime (30 μg), amikacin (30 μg) gentamicin (10 μg), imipenem (10 μg) and meropenem (10 μg). Clinical and Laboratory Standards Institute (CLSI) 2013 guidelines were used for interpretation.6
For the phenotypic detection of ESBL, combined disk method was used. Briefly, the test isolate was swabbed on Mueller Hinton agar plates after adjusting the opacity to 0.5 McFarland standard. Ceftazidime (30 μg) and ceftazidime/clavulanic acid (30/10 μg) discs was kept adjacent to each other at a distance of 20mm. After incubation at 37°C for 24 hrs, a zone size greater than 5mm with the ceftazidime/clavulanic acid disc when compared to the ceftazidime disc was considered as positive for ESBL production.7
The identification of P. aeruginosa by biochemical tests was confirmed with PCR. The boiling lysis method was used to extract DNA. An overnight culture was centrifuged at 10,000rpm for 10 minutes and to the supernatant 300μl of nuclease free water was added, boiled at 100°C for 10 minutes and stored at -20°C for at least 6 hours. The supernatant was collected after centrifugation at 10,000 rpm for 10 minutes and stored at 20°C. For PCR amplification, 2μl was used as a template and kept at -20°C for further use.8 PCR was performed for the confirmation of species using a species-specific primer which targets the 16S rRNA gene (Primer F: GGGGGATCTTCGGACCTCA and R: TCCTTAGAGTGCCCACCCG) and the amplicon size was 956bp.9 The presence of ESBL-encoding genes was determined using primer specific for blaVEB, blaPER, blaPSE, blaGES, blaTEM, blaSHV, blaCTX-M, blaBEL, blaOXA1, blaOXA10, blaOXA2 by simplex PCR (Table 1).2,10-11 For blaTEM positive isolates, IS26 transposon was detected by PCR.
Table (1):
Primers used for the detection of ESBL.
| Target gene | Sequence (5’ to 3’) | Expected amplicon size | Tm |
|---|---|---|---|
| VEB | F: ACGACTTCCATTTCCCATGC R: GGACTCTGCAACAAATACGC |
643bp | 58 |
| PER | F: AATGAATGTCATTATAAAAGC R: AATTTGGGCTTAGGGCAGAA |
925bp | 52 |
| GES | F: ATGCGCTTCATTCACGCAC R: CTATTTGTCCGTGCTCAGG |
846bp | 55 |
| OXA-1 | F: CAACGGATTAACAGAAGCATGGCTCG R: GCTGTRAATCCTGCACCAGTTTTCCC |
198bp | 60 |
| OXA-2 | F: GACCAAGATTTGCGATCAGCAATGCG R: CYTTGACCAAGCGCTGATGTTCYACC |
256bp | |
| OXA-10 | F: CGCCAGAGAAGTTGGCGAAGTAAG R: GAAACTCCACTTGATTAACTGCGG |
156bp | |
| CTX M-U | F: ATGTGCAGYACCAGTAARGT R: TGGGTRAARTARGTSACCAGA |
593bp | 50 |
| TEM | F: CATTTCCGTGTCGCCCTTATTC R: CGTTCATCCATAGTTGCCTGAC |
800bp | 60 |
| SHV | F: AAAGATCCACTATCGCCAGCAG R: ATTCAGTTCCGTTTCCCAGCGG |
231bp | |
| BEL | F: CGACAATGCCGCAGCTAACC R: CAGAAGCAATTAATAACGCCC |
600bp | 55 |
| PSE | F: AATGGCAATCAGCGCTTC R: GCGCGACTGTGATGTATA |
699bp |
The isolates were collected from inpatients and outpatients of all ages with wound and ear infections (pus 66%; n=142), followed by respiratory tract infections (sputum 15%; n=32, broncho alveolar lavage 2%; n=4, tracheal wash 1%; n=2), urinary tract infections (urine 10%; n=21), blood stream infections (blood 4%, n=8), other infections (semen 2%; n=4). Among the 213 P. aeruginosa isolates, highest antibiotic sensitivity was observed towards imipenem (90%) followed by cefepime (73%) and meropenem (72%). Highest resistance was observed towards ofloxacin (42%), gentamicin (38%) followed by piperacillin (37%) and ciprofloxacin (37%). Of the carbapenems tested, meropenem (25%) showed higher resistance than imipenem (8%) and a total of 22/213 (10%) isolates were resistant to both the carbapenems. Further, a total of 85/213(40%) and 58/213 (27%) isolates were resistant to third (ceftazidime) and fourth (cefepime) generation cephalosporins, respectively.
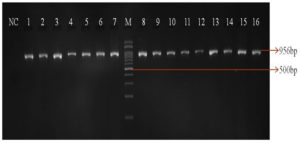
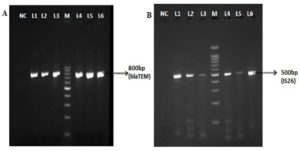
Of the 213 isolates, 85(40%) isolates were found to be resistant to ceftazidime and 27 (32%) isolates were confirmed as ESBL producing strains by the combined disk method. The remaining 58 isolates (68%) were resistant to the clavulanate combination, and this may be due to the overproduction of Amp C β-lactamases. All the tested isolates were confirmed as P. aeruginosa by species specific 16S rRNA PCR (Figure 1). PCR detected blaTEM in 44/85(52%) isolates and blaSHV gene was not identified in any of the isolates tested. Forty-four blaTEM positive isolates were further tested for IS26 transposon gene by PCR. Of the 44 isolates, 37 (84%) were positive for the IS26 transposase gene indicating their possible role in acquisition and mobilization of blaTEM (Figure 2).
Figure 2. Simplex PCR for detection of blaTEM and IS26. NC-Negative control; M-100bp ladder; Lane L1-L6 in the figure A and B represents the clinical isolates of P.aeruginosa showing positive for blaTEM and IS26 gene respectively
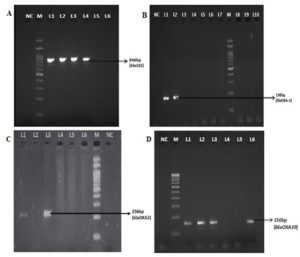
Among 85 clinical isolates, blaOXA-10 was detected in 19(23%) isolates; blaOXA-1, blaOXA-2, were found in 5(6%) and 3(4%) isolates respectively (Table 2, Figure 3). All blaOXA gene positive isolates exhibited the ESBL phenotype. blaVEB and blaPER types were found to be the most common ESBL in P. aeruginosa in several countries, whereas in this study all the isolates were negative for blaVEB and blaPER. The blaGES gene were identified in 4/82 (5%) isolates; two isolates showed resistant to both carbapenems tested; one isolate was susceptible to both the carbapenems, and another was resistant to meropenem and susceptible to imipenem. This shows varying extended activity of GES enzymes in hydrolyzing the carbapenem. All 4 blaGES positive isolates were sequenced and submitted to GenBank and accession numbers were obtained (Table 3). Among the Enterobacteriaceae, the most prevalent enzymes are the CTX-M group of enzymes; however, they were absent in this study. Other genes frequently seen in Pseudomonas such as blaVEB, blaPER and the minor ESBLs such as blaPSE and blaBEL were not found in any of the tested isolates.
Table (2):
Distribution of ESBL genes among P.aeruginosa.
Gene screened |
No. of positive isolates (%) |
|---|---|
VEB |
0 |
PER |
0 |
TEM |
44 (52%) |
SHV |
0 |
CTX M |
0 |
PSE |
0 |
BEL |
0 |
GES |
4 (5%) |
OXA – 1 |
5 (6%) |
OXA – 2 |
3 (4%) |
OXA -10 |
19 (22%) |
Table (3):
Sequencing of blaGES gene.
Strain id. |
Accession number |
|---|---|
PA34 |
MG696822 |
PAE136 |
MG589922 |
PAE129 |
MG742374 |
PAE140 |
MG755328 |
Figure 3. Simplex PCR for detection of blaGES, OXA-1, OXA-2 and OXA-10. NC-Negative control; M-100bp ladder; A: L1-L4 positive clinical isolates (blaGES), L5&L6 negative clinical isolates; B: L1&L2 positive clinical isolates (blaOXA-1), L3-L10 negative clinical isolates; C: L1 &L3 positive clinical isolates (blaOXA-2), L4-L6 negative clinical isolates; D: L1-L3 &L6 positive clinical isolates (blaOXA-10), L4 & L5 negative clinical isolates
Phenotypic detection of ESBL in P. aeruginosa is difficult due to the presence of various resistance mechanisms such as over expression of inducible chromosomal AmpC β-lactamase and greater degree of impermeability or efflux-mediated resistance.12 Current ESBL detection methods employed for Enterobacteriaceae are not recommended to be used for P. aeruginosa. Since there were no CLSI guidelines for P. aeruginosa in the year that this study was conducted, the interpretation criteria for Enterobacteriaceae was followed. Resistance to the clavulanate combination was observed in 58% of isolates and this may be due to the co-existence of ampC production or the presence of inhibitor resistant ESBL variants. The level of AmpC production may obstruct or even obscure the phenotypic detection. In ESBL-producing P. aeruginosa isolates, the spread of metallo beta lactamases may be another reason for the non-detection by phenotypic tests.4
PCR cannot differentiate the broad spectrum and extended spectrum variants of TEM β-lactamases. Previously, there have not been many reports of blaTEM and blaSHV genes in P. aeruginosa, due to the high prevalence of oxacillinase and carbenicillinase genes, upregulation of chromosome-encoded cephalosporinase and the rarity of narrow-spectrum TEM type enzymes.2 In this study, blaTEM was the most prevalent followed by blaOXA-10 whereas another study showed that the most prevalent ESBL gene was blaVEB followed by blaTEM, blaGES and blaSHV.13 These may vary in different geographical regions.
Our study result shows that blaTEM was found in 52% of isolates and blaSHV was not detected in any of the isolates. The study further demonstrated the presence of IS26 association with blaTEM gene. Horizontal gene transfer mechanisms may have played a role in the dissemination of this kind of enzymes from the Enterobacteriaceae family. The selective pressure of antibiotics on the bacteria could have modified the resistant mechanisms.14 CTX-M variant was previously one of the most frequently reported enzymes in Enterobacteriaceae; later it was reported in P. aeruginosa.15 Dissemination of the gene may be due to the insertion sequence ISEcp1 which is frequently observed upstream of the blaCTX-M genes. However, in this study, none of the isolates exhibited blaCTX-M which differed from previous findings.16-19
Molecular class D β-lactamases mainly OXA group exhibit greater diversity in their enzymatic activities and are the most frequent ESBLs found in Pseudomonas sp. The present study reports the presence of blaOXA-10, blaOXA-1 and blaOXA-2 in 19 isolates, 5 isolates and 3 isolates respectively. OXA-10 and to a lesser extent, OXA-2 are the origin of the majority of OXA-type ESBLs.20 OXA 10 and OXA 2 are classified as narrow-spectrum Class D β-lactamases; however, when expressed in A. baumannii, they can show carbapenemase activity.21 In the present study, blaGES was identified in four isolates which showed both the ESBL phenotype and carbapenemase activities. This could be a stage in the transition between ESBLs and carbapenem-hydrolyzing enzymes of class A. GES has previously been observed in different places in India.22 The high frequency of ESBLs in the present study indicates the necessity for standardization of phenotypic method of detection and increasing its sensitivity. Further, PCR is essential for the characterization of ESBLs and monitoring the dissemination of resistant genes. Although the prevalence of ESBLs has been documented in numerous studies from around the world, geographical and institutional differences in their prevalence have been observed.
ESBL-producing P. aeruginosa strains are reported world-wide. Use of molecular methods for the detection of resistant genes is important to study their prevalence.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both the authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was funded by University Grant Commission, New Delhi, India with reference No: F19-121/2014(BSR).
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Human Ethics Committee, Dr. ALM PG IBMS, University of Madras, Chennai, India with reference number IHEC no:UM/IHEC/02-2014-I.
- Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45(6):568-585.
Crossref - Weldhagen GF, Poirel L, Nordmann P. Ambler Class A Extended-Spectrum β-Lactamases in Pseudomonas aeruginosa: Novel Developments and Clinical Impact. Antimicrob Agents Chemother. 2003;47(8):2385-2392.
Crossref - He S, Hickman AB, Varani AM, et al. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio. 2015;6(3):e00762-15.
Crossref - Laudy AE, Rog P, Smolinska-Krol K, et al. Prevalence of ESBL-producing Pseudomonas aeruginosa isolates in Warsaw, Poland, detected by various phenotypic and genotypic methods. PLoS One. 2017;12(6):e0180121.
Crossref - Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol. 1966;45(4):493-496.
Crossref - CLSI. M100-S24 Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. 2013.
- Taneja N, Sharma M. ESBLs detection in clinical microbiology: why & how? Indian J Med Res. 2008;127(4):297-300. https://pubmed.ncbi.nlm.nih.gov/18577783/.
- Pitout JDD, Gregson DB, Poirel L, McClure JA, Le P, Church DL. Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43(7):3129-3135.
Crossref - Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42(5):2074-2079.
Crossref - Strateva T, Ouzounova-Raykova V, Markova B, Todorova A, Marteva-Proevska Y, Mitov I. Problematic clinical isolates of Pseudomonas aeruginosa from the university hospitals in Sofia, Bulgaria: current status of antimicrobial resistance and prevailing resistance mechanisms. J Med Microbiol. 2007;56(7):956-963.
Crossref - Voets GM, Fluit AC, Scharringa J, Stuart JC, Leverstein-van Hall MA. A set of multiplex PCRs for genotypic detection of extended-spectrum β-lactamases, carbapenemases, plasmid-mediated AmpC β-lactamases and OXA β-lactamases. Int J Antimicrob Agents. 2011;37(4):356-359.
Crossref - Jiang X, Yu T, Jiang X, Zhang W, Zhang L, Ma J. Emergence of plasmid-mediated quinolone resistance genes in clinical isolates of Acinetobacter baumannii and Pseudomonas aeruginosa in Henan, China. Diagn Microbiol Infect Dis. 2014;79(3):381-383.
Crossref - Pragasam AK, Vijayakumar S, Bakthavatchalam YD, et al. Molecular characterisation of antimicrobial resistance in Pseudomonas aeruginosa and Acinetobacter baumannii during 2014 and 2015 collected across India. Indian J Med Microbiol. 2016;34(4):433-441.
Crossref - Lerminiaux NA, Cameron AD. Horizontal transfer of antibiotic resistance genes in clinical environments.Can J Infect Dis. 2019;65(1):34-44.
Crossref - Al Naiemi N, Duim B, Bart A. A CTX-M extended-spectrum β-lactamase in Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J Med Microbiol. 2006;55(11):1607-1608.
Crossref - Picao RC, Poirel L, Gales AC, Nordmann P. Further identification of CTX-M-2 extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(5):2225-2226.
Crossref - Polotto M, Casella T, de Lucca Oliveira MG, et al. Detection of P. aeruginosa harboring bla CTX-M-2, blaGES-1 and blaGES-5, bla IMP-1 and bla SPM-1 causing infections in Brazilian tertiary-care hospital. BMC Infect Dis. 2012;12(1):176.
Crossref - Saxena S, Banerjee G, Garg R, Singh M. CTX-M and PER-1 group extended spectrum β-lactamases-producing Pseudomonas aeruginosa from the patients of lower respiratory tract infection. Indian J Med Microbiol. 2015;33(1):191-192.
Crossref - Gupta R, Malik A, Rizvi M, Ahmed M. Presence of metallo-beta-lactamases (MBL), extended-spectrum beta-lactamase (ESBL) & AmpC positive non-fermenting Gram-negative bacilli among Intensive Care Unit patients with special reference to molecular detection of blaCTX-M & blaAmpC genes. Indian J Med Res. 2016;144(2):271.
Crossref - Naas T, Poirel L, Nordmann P. Minor extended-spectrum β-lactamases. Clin Microbiol Infect. 2008;14:42-52.
Crossref - Antunes NT, Lamoureaux TL, Toth M, Stewart NK, Frase H, Vakulenko SB. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother 2014;58(4):2119-2125.
Crossref - Maurya AP, Choudhury D, Talukdar AD, Dhar A, Chakravarty A, Bhattacharjee A. A report on the presence of GES-5 extended spectrum beta-lactamase producing Pseudomonas aeruginosa associated with urinary tract infection from north-east India. Indian J Med Res. 2014;140(4):565-567. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4277147/
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.