ISSN: 0973-7510
E-ISSN: 2581-690X
The current study was aimed to detect the existence of genes encode outer membrane proteins: transfer protein (traT) and increased serum survival (iss ) which associated with resistance to complement bacterial lysis activity in different Enterobacteriaceae species isolates .Enterobacteriaceae isolates included in this study were (5) isolates of E. coli , K. pneumonia, Salmonella typhi and Shigella dysentery, (4) isolates of Proteus vulgaris and (2) isolates of Serratia marcescens which were isolated from different clinical infections . The traT and iss genes were revealed from whole DNA of 26 isolates of Enterobacteriaceae family, it was found that 21(80.7%) of Enterobacteriaceae isolates gave positive result for traT gene at 288bp while only 17(65.3%) of isolates gave positive result for iss gene at 258bp. It was observed that the traT gene was recognized among 100 % of Salmonella typhi and Serratia marcescens strains and the prevalence of a traT gene was showed among 80 % of K. pneumoniae and Shigella dysentery isolates whereas 75% of Proteus vulgaris and 60% of E. coli isolates contain the traT gene. The iss was found among 80% of both E. coli and Shigella dysentery isolates while it was observed in 60% of K. pneumonia and Salmonella typhi isolates and it was present only in 50% of Proteus vulgaris and Serratia marcescens isolates, as conclusion the study confirmed presence of traT and iss which linked to human complement resistance among local Enterobacteriaceae species and high rate of occurrence among extraintestinal isolates.
Enterobacteriaceae, PCR, complement resistance, traT, and iss
The Enterobacteriaceae family contains a large number of species that are causes for most common infections like nosocomial infections, urinary tract and wound infections, pneumonia, meningitis and septicemia (Ruiz et al., 2002). The pathogenicity of this group is due to several factors include adhesins, hemolysin secretion, serum resistance and biofilm creation . Also, it is probable that the genotypes and phylogenetic origin of Enterobacteriaceae varies according to geographical areas (Johnson et al., 2002 and Martý´nez et al., 2006). Outer membrane proteins (OMPs) are a group of proteins exist in the outer membrane of Gram-negative bacteria. These proteins preserve the bacteria in a aggressive environment and also assistance in a number of activities including moving of solute through the impassable outer membrane and signal transduction .These proteins are also complicated in connection and invasion of the bacteria (Koebnik et al., 2000). OMPs have different functions in the activation of the immune system of the host, such as resistance against antimicrobial peptides and activation of dendritic cells (Jeannin et al.,2000). Resistance to the bactericidal action of complement is a distinctive of all gram-negative bacteria that cause septicemia (Joiner, 1988). OMPs of some Gram-negative strains are possibly related with the sensitivity to the bactericidal action of complement (Futoma-Koloch et al., 2006). Previously the role of OMPs in resistance to bactericidal activity of complement was confirmed in S. typhimurium and Y enterocolitica, K. pneumoniae P. mirabilis (Heffernan et al.,1994, Alberti et al.,1996 and Kaca et al.,2009). The bactericidal activity of complement is acquired by the transfer protein called traT which was showed in other studies in E. coli, Salmonella, Shigella and Klebsiella strains (Montenegro et al., 1985, Fernandez-Beros et al., 1990 and Wu et al., 2007).
The increased serum survival protein (iss) has an action in defense to serum complement (Nolan et al.,2003). The iss has been known for its activity in virulence of extraintestinal pathogenic Escherichia coli strains (ExPEC) (Hassan, 2011). The iss gene is recognized as one of the most common virulence genes in extraintestinal pathogenic strains in poultry (Badouei et al., 2015). The occurrence of immune resistance among Enterobacteriaceae family was a cause to study the prevalence of genes associated with serum resistance among Enterobacteriaceae isolates in Babylon province as an indicator for appearance of resistance of Enterobacteriaceae infections .
Isolation and Identification
Twenty-six clinical isolate of Enterobacteriaceae were collected from different clinical sources (urine , blood, stool and sputum) from patients in Al-Hilla teaching hospital /Babylon province during the period from November 2017 to March 2018. Specimens were cultured on selective media and recognized by biochemical tests as mentioned by forbes et al., 2007. Identification was confirmed by the automated method Vitek-2 system (BioMérieux, France).
Genetic detection of Virulence genes
The conventional PCR technique was used for recognition of virulence genes include (traT and iss).
Extraction of Bacterial DNA
The whole bacterial DNA was extracted from bacterial isolates by using Genomic DNA Mini Bacteria Kit that provided by the company (Geneaid, UK). The DNA solution was stored at -20°C till used in PCR.
PCR Amplifications
Recognition of virulence genes in E. coli ,Klebsiella pneumonia, Shigella dysenteriae, Serratia marcescens, Salmonella typhi, Proteus vulgaris isolates was achieved by amplifying conventional PCR. The PCR primers were provided by (Macrogen company, Korea). The product size and sequences of the PCR primers are showed in Table 1.
The distribution of bacterial genes according to the site of infection
A total of 26 Enterobacteriaceae isolates were isolated from different clinical cases and were diagnosed by using vitek -2 system , the number each isolates were (5) E.coli, (5) K.pneumoniae, (5) Salmonella typhi, (5) Shigella dysentery, (2) Serratia marcescens and (4) Proteus vulgaris. The prevalence of traT and iss genes was 100% in both of respiratory and intestinal isolates while the percent of traT and iss genes in blood isolates were 100% and 50%, respectively whereas the prevalence of virulence genes in blood culture isolates which have (68.7%) and the occurrence of traT and iss genes observed in UTI isolates was 80 % and 60%, respectively (Table 1). The virulence-associated phenotypes including complement resistance, adhesion and invasion, and maintenance within macrophages in S. typhimurium have been caused by different members of OMPs group (Pulkkinen and Miller ,1991 and Heffernan et al., 1994). The contribution of OMPs has been showed in bacterial serum-resistance by their inactivation of complement at the C3, C9, and C5b-9 steps (Wooley et al., 1993 and Biedzka-sarek et al., 2005). The iss gene was first recognized in a human septicemic E. coli strain (Nolan et al.,1992). The iss and traT genes were present among 95.5% and 86.4% of E.coli strains of blood infections and only among 68.8 % and 43.8% of the intestinal infections , respectively (Fernandez-Beros et al., 1990), this result is comparable with the current study. The occurrence of the iss gene was higher in tissue isolates that clarify the role of this protein in pathogenesis of a vain pathogenic E.coli APEC strains (Nolan et al., 1992 and Johnson et al., 2008a). The traT genes were existent in most of the ETEC E.coli strains, this is may be due to ColV plasmids that are capable to transmission fully among varied E. coli strains, giving these strains some virulence-related characteristics (Fernandez-Beros et al., 1990).
Table (1):
The size and sequences of primers used in current study.
| References | size (bp) | Primer Sequence | Gene name | |
|---|---|---|---|---|
| ( Hassan ,2011) | 258 | GGCAATGCTTATTACAGGATGTGC GAGCAATATACCCGGGCTTCC |
F R |
Iss |
| ( El Fertas-Aissani ,et al.,2013) | 288 | GGTGTGGTGCGATGAGCACAG CACGGTTCAGCCATCCCTGAG |
F R |
traT |
Molecular detection of virulence factors genes among Enterobacteriaceae isolates
The traT and iss genes which encoded important virulence factors that are linked with human serum resistance were genetically scanned by using PCR technique for recognition of the existence of these genes in 26 isolates of Enterobacteriaceae family, it was found that 21(80.7%) of Enterobacterial isolates were contained traT gene at 288bp while only 17(65.38%) of isolates were contained iss gene at 258bp. A group of outer membrane protein (OMP) genes has been documented in the Enterobacteriaceae species including Escherichia coli, Shigella flexneri, Salmonella typhimurium, Klebsiella pneumoniae, Serratia marcescens, Proteus vulgaris, Proteus mirabilis, and Providencia stuartii. One or two peptidoglycan-associated main outer membrane proteins were found in all studied strains (Hofstra et al.,1980). The current data approved with other study which detect the commonness of traT in Gram negative bacteria, particularly E. coli, Salmonella, Shigella and Klebsiella, It was showed in a higher percentage among the E. coli strains isolated from the blood stream of patients with bacteremia /septicemia or from feces of patients with gastrointestinal infections (50-70%) while the incidence of traT in strains isolated from cases of urinary tract infections was variable (Montenegro et al.,1985).
Table (2):
The dissemination of genes among bacterial isolates according to the site of infection.
Source of isolates |
traTn (%) |
issn (%) |
|---|---|---|
Respiratory infections |
2/2(100%) |
2/2(100%) |
Intestinal infections |
7/10(70%) |
7/10(70%) |
Blood |
4/4(100%) |
2/4(50%) |
Urinary tract infections |
8/10(80%) |
6/10(60%) |
Total |
21/26(80.7%) |
17/26(65.3%) |
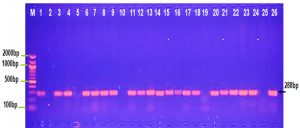
The PCR result observed in Figure 1 shows that 100 % of Salmonella typhi and Serratia marcescens isolates gave positive result for traT gene. OMPs has significant role in pathogenesis of S. typhimurium and Y enterocolitica (Heffernan et al.,1994).
Table (3):
The occurrence of virulence factors genes among bacterial species.
Species |
Number |
traT n (%) |
iss n (%) |
|---|---|---|---|
E.coli |
5 |
3/5(60%) |
4/5(80%) |
Klebsiella pneumonia |
5 |
4/5(80%) |
3/5(60%) |
Salmonella typhi |
5 |
5/5(100%) |
3/(60%) |
Shigella dysentery |
5 |
4/5(80%) |
4/5(80%) |
Proteus vulgaris |
4 |
3/4(75%) |
2/4(50%) |
Serratia marcescens |
2 |
2/2(100%) |
1/2(50%) |
Total |
26 |
21/26(80.7%) |
17/26(65.38%) |
In the current study traT gene was identified in 80% K. pneumoniae and Shigella dysentery isolates, this result compatible with other study which found that traT gene was detected in (78.5%). of K. pneumoniae (Wasfi et al., 2016) while Kuo et al., 2017 (Kus et al., 2017) detected traT gene in 11.3% of K.pneumonia isolates. 75% and 60% of E. coli and Proteus isolates were positive for traT gene, respectively (Fig. 1), this result parallel with other study by Wooly et al., 1993 who showed that 57.1% of E.coli isolates were contained traT gene and Wu et al., 2007 showed presence of traT genes in most of the E.coli O149 isolates. Fernandez-Beros et al.,1990 showed that the iss and traT genes were existent among 95.5 % and 86.4 % of the E.coli blood isolates and only among 68.8 % and 43.8 % of the intestinal isolates, correspondingly. Previously it was described a ColV plasmid, among E.coli isolates from blood and intestinal sources, ColV plasmids confer Escherichia coli with some virulence genes including iss and traT (Aguero et al., 1984 and Skyberg et al.,2008). ColV plasmids are capable to transmission completely among diverse E. coli strains, providing those strains with several virulence-linked features (Fernandez-Beros et al., 1990).
Fig. 1. Agarose gel electrophoresis image that shown the PCR product of a virulence factor gene (traT) at (258bp) in different bacterial isolates. Where M: Marker (2000-100bp), lane (1-5) E. coli, lane (6-10) K. pneumonia , lane (11-15) Salmonella typhi, lane (16-20) Shigella dysentery , lane (21-22) Serratia marcescens and lane (23-26) Proteus vulgaris.
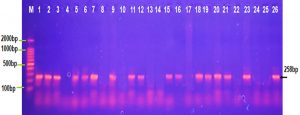
The occurrence of iss gene product at 258 bp in E. coli and Shigella dysentery isolates was 80% while it was 60% among K. pneumonia ,Salmonella typhi isolates and 50% of Proteus vulgaris and Serratia marcescens isolates as showed in Fig. 2. This result comparable with a study of Badouei et al., 2015 who found that the occurrence of the iss gene in a vain E.coli strains was 90.3% in the septicemic isolates and 64.3% among cecal isolates and also approved with a study of Vaz et al., 2017 who observed the occurrence of iss gene was ranged from 75 to 91% among the ExPEC strains and was ranged from 49 to 57% among the fecal strains ,while other study showed that 30% of uropathogenic E.coli isolates were positive for iss gene and only 8% of K.pneumoniae isolates were contained iss gene (Hassan, 2015).
Fig. 2. Agarose gel electrophoresis image that shown the PCR product of a virulence factor gene (iss) at (258 bp) in different bacterial isolates. Where M: Marker (2000-100bp), lane (1-5) E. coli, lane (6-10) K. pneumonia , lane (11-15) Salmonella typhi, lane (16-20) Shigella dysentery , lane (21-22) Serratia marcescens and lane (23-26) Proteus vulgaris
The iss gene was represented one of the most prevalent virulence genes in extra-intestinal pathogenic strains and three alleles of iss were recognized among E. coli isolates (Johnson et al., 2008b). Human serum resistance among ExPEC permits persistence in the host’s blood stream and can possibly distribute to cause human infections through the food chain is indistinct, it can colonize the intestinal area and prompt virulence gene transmission to human strains. For example, APEC strains and urogenital pathogenic E. coli (UPEC) causing human infections were revealed to share large genomic sequences (Johnson et al.,2008a and Kariyawasam et al.,2007).
The present study concluded that high distribution of genes encoded for outer membrane proteins responsible for human complement resistance among Enterobacteriaceae species isolated from different clinical infections and also these genes were detected among extraintestinal infections isolates .
- Aguero, M. E. et al. A plasmid-encoded outer membrane protein, TraT, enhances resistance of Escherichia coli to Phagocytosis, Infect. Immun., 1984; pp. 740-746.
- Alberti, S. et al. Analysis of complement C3 deposition and degradation on Klebsiella pneumoniae. Infect. Immun. 1996; 64:4726–4732.
- Badouei,A. M. et al. ‘Prevalence and clonal distribution of avian Escherichia coli isolates harboring increased serum survival (iss) gene’, Journal of Applied Poultry Research, 2015; 25(1), pp. 67–73. doi: 10.3382/japr/pfv064.
- Biedzka-sarek, M., Venho, R. and Skurnik, M. ‘Role of YadA , Ail , and Lipopolysaccharide in Serum Resistance of Yersinia enterocolitica Serotype O/ : 3 Role of YadA , Ail, and Lipopolysaccharide in Serum Resistance of Yersinia enterocolitica Serotype O/ : 3’, 2005; 73(4), pp. 2232–2244. doi: 10.1128/IAI.73.4.2232.
- El Fertas-Aissani, R. et al. ‘Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens.’, Pathologie-biologie, 2013; 209–16. doi: 10.1016/j.patbio.2012.10.004.
- Fernandez-Beros, M.E. et al. ‘Virulence-Related Genes in Co1V Plasmids of Escherichia coli Isolated from Human Blood and Intestines, 1990; 28(4), 742–746.
- Forbes, B. A., Daniel, F S. and Alice, S. W. Baily and Scot s Diagnostic microbiology.12th Mos, USA, U, 2007.
- Futoma-Koloch, et al. Survival of Proteus mirabilis O3 (S1959), O9 and O18 strains in normal human serum (NHS) correlates with the diversity of their outer membrane proteins (OMPs). Pol J Microbiol., 2006; 55, 153–156.
- Hassan, R. ‘Characterization of Some Virulence Factors Associated with Enterbacteriaceae Isolated From Urinary Tract Infections in Mansoura Hospitals’, Egyptian Journal of Medical Microbiology, 2011; 20(2), 9–17.
- Heffernan, J.E.; Wu, L.; Louie,J.; Okamoto, S.; Fierer, J. and Guiney,D.J. ‘Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella typhimurium and ail from Yersinia enterocolitica’, Infection and Immunity, 1994; 62(11), 5183–5186.
- Hofstra, H., Van Tol, M.J.D. and Dankert, J. ‘Cross-reactivity of major outer membrane proteins of Enterobacteriaceae, studied by crossed immunoelectrophoresis’, Journal of Bacteriology, 1980; 143(1), 328–337.
- Jeannin, P. et al. OmpA targets dendritic cells, induces their maturation and delivers antigen into the MHC class I presentation pathway. Nat. Immunol., 2000; 1, 502–509.
- Johnson, J. R. et al. Epidemiological correlates of virulence genotype and phylogenetic background among Escherichia coli blood isolates from adults with diverse-source bacteremia. J Infect Dis., 2002; 185, 1439–1447.
- Johnson, T. J. et al. ‘Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool’, Journal of Clinical Microbiology, 2008a; 46(12), 3987–3996. doi: 10.1128/JCM.00816-08.
- Johnson, T.J., Wannemuehler, Y. M. and Nolan, L. K. ‘Evolution of the iss gene in Escherichia coli, Applied and Environmental Microbiology, 2008b; 74(8), 2360–2369. doi: 10.1128/AEM.02634-07.
- Joiner, K. A. Complement evasion by bacteria and parasites. Annu. Rev. Microbiol., 1988; 42:201-230.
- Kaca, W. et al. ‘Human complement activation by smooth and rough Proteus mirabilis lipopolysaccharides’, Archivum Immunologiae et Therapiae Experimentalis, 2009; 57(5), 383–391. doi: 10.1007/s00005-009-0043.
- Kariyawasam, S., Scaccianoce, J. A. and Nolan, L.K. ‘Common and specific genomic sequences of avian and human extraintestinal pathogenic Escherichia coli as determined by genomic subtractive hybridization’, BMC Microbiology, 2007; 7, 1–8. doi: 10.1186/1471-2180-7-81.
- Koebnik, R., Locher, K. P. and Van Gelder, P. ‘Structure and function of bacterial outer membrane proteins: barrels in a nutshell.’, Molecular microbiology, 2000; 37(2), 239–253. doi: 10.1046/j.1365-2958.2000.01983.x.
- Kuº , H.; Arslan, U.; Türk, D. H and Fýndýk, D. Investigation of various virulence factors of Klebsiella pneumoniae strains isolated from nosocomial infections. Mikrobiyol Bul. 2017; 51(4):329-339.
- Martý´nez, J. A.et al.. Relationship of phylogenetic background, biofilm production, and time to detection of growth in blood culture vials with clinical variables and prognosis associated with Escherichia coli bacteremia. J. Clin. Microbiol., 2006; 44,pp. 1468–1474.
- Montenegro, M. et al. ‘traT gene sequences, serum resistance and pathogenicity-related factors in clinical isolates of Escherichia coli and other gram-negative bacteria.’, Journal of general microbiology, 1985; 131(6), 1511–1521. doi: 10.1099/00221287-131-6-1511.
- Nolan, L. K., R. E Wooley, and R. K. Cooper. Transposon mutagenesis used to study the role of complement resistance in the virulence of an avian Escherichia coli isolate. Avian Dis., 1992; 36, 398–402.
- Nolan, L. K., S. M. Horne, C. W. Giddings, S. L. Foley, T. J. Johnson, A. M. Lynne, and J. Skyberg. Resistance to serum complement, iss, and virulence of avian Escherichia coli. Vet. Res. Commun. 2003; 27:101–110.
- Pulkkinen, W. S., and S. I. Miller. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocoitica invasion protein and a bacteriophage lambda outer membrane protein. J. Bacteriol., 1991; 173,pp.86-93.
- Ruiz, J. et al. Differences in virulence factors among clinical isolates of Escherichia coli causing cystitis and pyelonephritis in women and prostatitis in men. J. Clin. Microbiol., 2002; 40, 4445–4449.
- Skyberg, J.A.; Johnson, T.J.and Nolan, L.K. Mutational and transcriptional analyses of an avian pathogenic Escherichia coli ColV plasmid. BMC Microbiol., 2008; 29;8:24.
- Vaz, R. V. et al. ‘Phylogenetic characterization of serum plus antibiotic-resistant extraintestinal Escherichia coli obtained from the liver of poultry’, 2017; 37(10), 1069–1073. doi: 10.1590/S0100-736X2017001000005.
- Wasfi, R., Elkhatib, W. F. and Ashour, H. M. ‘Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals’, Scientific Reports. Nature Publishing Group, 2016; 6, 1–11. doi: 10.1038/srep38929.
- Wooley,R.E. et al. Association of K-1 Capsule, Smooth Lipopolysaccharides, traT Gene, and Colicin V Production with Complement Resistance and Virulence of Avian Escherichia coli. Avain disease, 1993; 37, 1092-1096 .
- Wu, X. et al. ‘Comparative Analysis of Virulence Genes , Genetic Diversity , and Phylogeny of Commensal and Enterotoxigenic Escherichia coli Isolates from Weaned Pigs’, 2007; 73(1), 83–91. doi: 10.1128/AEM.00990.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.