ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.1.28 | © The Author(s). 2019
Staphylococcus aureus, including methicillin resistant S. aureus (MRSA) is the most commonly isolated pathogen in hospitals worldwide. The aim of present study was molecular characterization of Staphylococcus aureus isolated from renal hemodialysis (HD) patients from Ha’il region of Saudi Arabia. A total of 392 samples were screened from 204 HD patients for colonization of S. aureus. The isolated bacteria were identified by MALDI-TOF-MS. Antibiotic susceptibility testing was performed using Microscan. Among these isolates, 72 S. aureus (43% MRSA and 57% MSSA) were identified.The isolates were considerably resistant with varied profile to the antibiotics tested except being 100% susceptible to vancomycin, linezolid and teicoplanin. Of the isolates, 22.2% were positive for biofilm assay. Four representative MRSA isolates were selected and whole genome sequence analysis was performed using MiSeq. Two out of the 4 MRSA were found to be ST-1 and 2 were found to be ST-32. Among MRSA isolates, 25.8% were negative for mecA and all of them were negative for mecC gene. A high prevalence of MRSA in HD patients as well as high percentage of biofilm production in MRSA isolates highlights the vital role for standardized surveillance along with validated molecular typing methods to evaluate the incidence of MRSA and accordingly to control its spread.
Hemodialysis, Staphylococcus aureus, Whole genome sequencing Pathogenic bacteria, MALDI- TOF-MS.
Staphylococcus aureus is an opportunistic bacterial pathogen responsible for a large number of human and animal infections. Staphylococcus aureus is associated with asymptomatic colonization of the skin and mucosal surfaces of about 30% of normal humans1,2. The Staphylococcal infections have been found regularly among the patients with compromised immune system and when the skin or mucosal barriers are breached, following insertion of a foreign body3.
There is a high incidence of infections caused by S. aureus among the patients with renal disease; specifically, those undergoing hemo-dialysis or kidney transplantation4,5. Because of frequent use of antimicrobials for a prolonged time and use of catheters, the hemodialysis (HD) patients are at a higher risk of colonization and infection by multi-drug resistant S. aureus including MRSA (methicillin resistant S. aureus)6-8. The bacterial infections are the major cause of morbidity and mortality during receiving hemodialysis and S. aureus, particularly MRSA, is one of the most common pathogen9-11. Mortality from all causes in patients on dialysis treatment is 6.5–7.9 times higher than that of the general population. S. aureus can colonize almost half the dialysis population without any indication of disease12. However, such colonization of S. aureus can cause wound and tissue infections; septicemia; and chronic infections13-16.
Infections cause significant morbidity and mortality among dialysis patients and HD patients are a high-risk population for blood-stream infection17,18. Renal disease, especially hemodialysis is a complex health care issue globally, including Saudi Arabia. Saudi Center for Organ Transplantation (SCOT) estimated a total of 19,659 dialysis patients, 18,270 of them are treated by hemodialysis (HD) and the remaining 1,389 by peritoneal dialysis (PD) with the mortality of about 9%19. There is a lack of data regarding the prevalence of S. aureus among hemodialysis patients from Saudi Arabia; therefore, the aim of present study was molecular characterization of Staphylococcus aureus isolated from renal hemodialysis (HD) patients from Ha’il region of Saudi Arabia.
Bacterial isolates
In this study, a total of 392 samples were screened from 204 HD patients from King Khalid Hospital, Ha’il, Saudi Arabia. The samples were collected from catheter tips, catheter site swabs and nose swabs.TOF-MS
The identification of bacterial isolates was performed on MALDI-TOF-MS (Bruker Daltonics Germany) according to the manufacturer’s guidelines. Briefly, a fresh bacterial colony from overnight culture was smeared on target plate overlaid with 1 ml of a saturated a-cyano-4-hydroxy-cinnamic acid (HCCA) matrix solution in 50% acetonitrile-2.5% trifluoroacetic acid (Bruker Daltonics) with the help of a sterile toothpick and air dried at room temperature. The plate was loaded in to the machine and the operation was run. The identification and analysis of mass spectra were performed the MALDI Biotyper software package (version 3.0).
Identification and Antibiotic susceptibility by Microscan
Microscan walkaway (Siemens Healthcare Diagnostics, Sacramento, CA, USA); an automated system used for bacterial identification and antibiotic susceptibility test was used for confirmation of identification and antimicrobial susceptibility of the bacterial strains. In this method, a small portion of a well isolated colony was taken and added to a Gram-positive Microscan combo panel. The panel was loaded into the Microscan walkaway machine according to the manufacturer’s protocol. Results were available after 24- 48 hrs.
Biofilm assay
Biofilm assay was performed according to a previously published method20.
Polymerase Chain Reaction for mec genes
PCR was used to determine the type of mec gene in MRSA isolate21. The primers used for detection of types of mec gene were F: 52 -GTAGAAATGACTGAACGTCCGATGA-32 and R: 52 -CCAATTCCACATTGTTTCGGTCTAA-32 .
A PCR amplicon of 310 base pairs was analyzed using Sanger sequencer.
Whole Genome Sequencing
The sequencing of the bacterial genome for detection of antibiotic resistant genes, virulence factors, plasmids and MLST types was performed by using Illumina methodology using NextEra kit for library preparation22. The presence of known acquired resistance genes was determined by mapping the data from the isolate to an online database. The ResFinder web server (www.genomicepidemiology.org) and Basespace from Illumina was used to identify acquired antimicrobial resistance genes, MLST types and the presence of different virulent genes in the WGS data, using a threshold of 98% identity.
There is a high incidence of colonization followed by infections caused by S. aureus among the HD patients5. The main reason behind this high infection rate among HD patients is because of frequent use of antimicrobials for a prolonged time and use of catheters during the dialysis procedure. Thus HD patients are at a higher risk of colonization and infection by multi-drug resistant S. aureus including MRSA (methicillin resistant S. aureus)6-8. The bacterial infections are the major cause of morbidity and mortality during receiving hemodialysis and S. aureus, particularly MRSA, is one of the most common pathogen9-11. The current study was aimed at characterization of S. aureus isolated from renal hemodialysis (HD) patients from Ha’il region of Saudi Arabia.
A total of 72 S. aureus isolates were cultured from patients undergoing HD, and among these, 43.1% were MRSA and 56.9% were methicillin sensitive S. aureus (MSSA). Previous studies have highlighted a high percentage of S. aureus colonization among HD patients17,23,24. The percentage of MRSA and MSSA in our study was found similar to that of a study published from Japan17. The antibiotic profiling of S. aureus is very critical in management of the serious infections among the hospitalised patients. The S. aureus isolates from our study showed a varied profile to the antibiotics tested with 100% susceptibility to vancomycin, linezolid and teicoplanin. In addition, the capability of biofilm production among S. aureus helps it to remain in the hospital environment for prolonged time period leading to colonization of more patients25. In our study, 22.2% S. aureus were positive for biofilm assay.
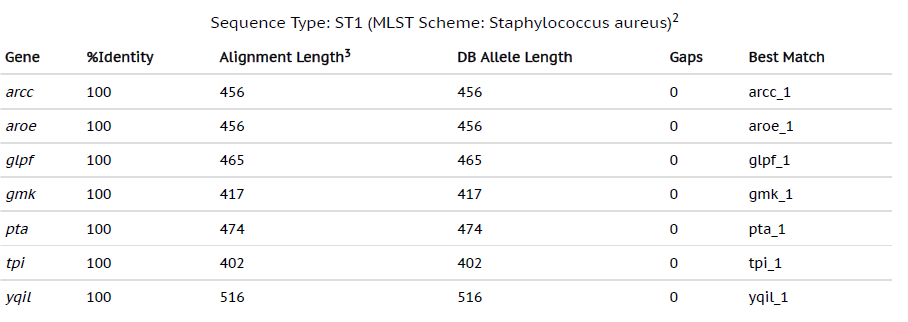
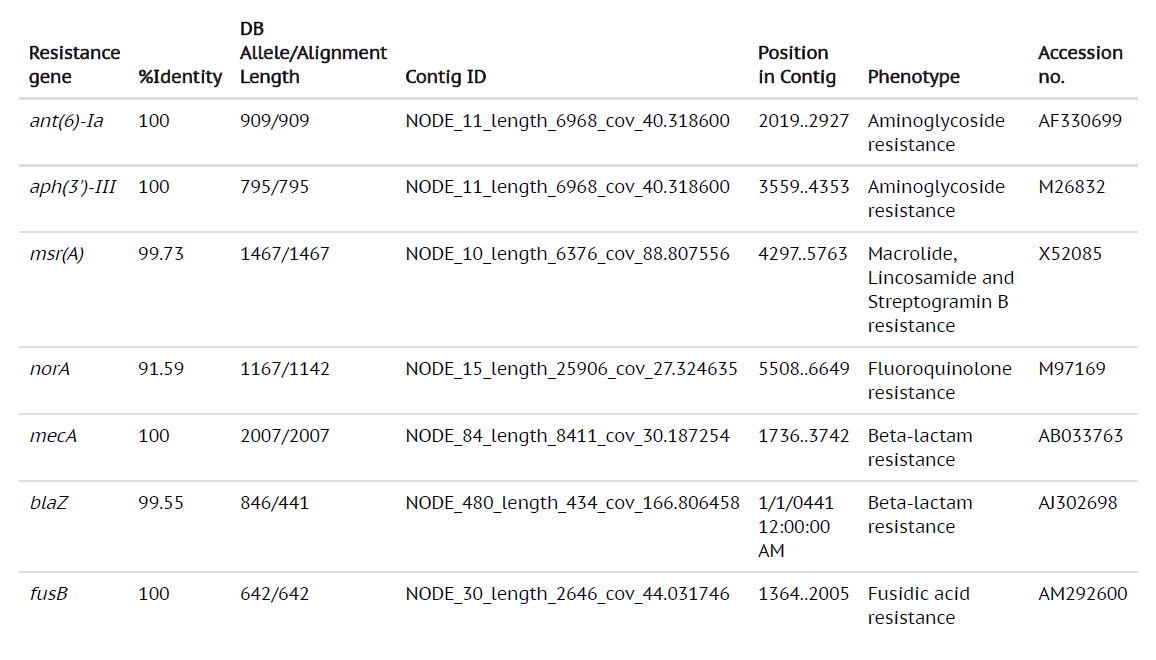
By using the most advanced technique in Microbiology laboratory, whole genome sequencing can provide a broad analysis of the bacterial strains from all the sources. With the development of bench-top sequencers and rapid analytical softwares, WGS has become a useful tool to guide treatment of the infections caused by bacterial strains. The whole genome sequencing results of MRSA from our study revealed that genome sizes ranging from 2879711 bp to 3012628 bp with 720 to 3408 contigs were successfully sequenced. The MLST data revealed that the most common MLST type of the MRSA from our study were ST1 and ST80 (Table 1 and Table 2). The res finder showed that the MRSA in our study contained the genes which exhibited the resistance to aminoglycosides, macrolides, fluoroquinolones, b-lactams and the most common genes detected were msr(A), norA, blaZ, ant(6)Ia, aph(3)-III, mecA and fusB (Table 3). The presence of these genes by WGS was found to be associated with that of phenotypic antibiotic profiles.
Table (1):
Whole genome sequencing results showing MLST type 1
Table (2):
Whole genome sequencing results showing MLST type 80
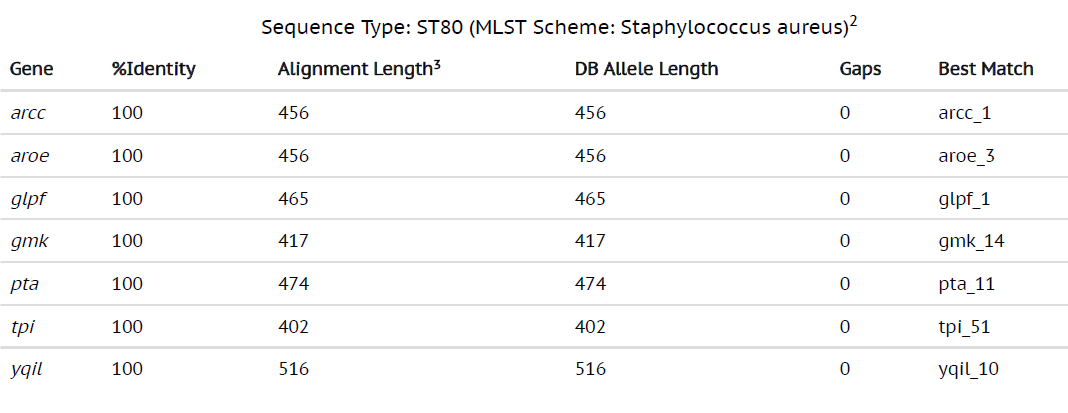
Table (3):
Whole genome sequencing results showing different antibiotic resistance genes and virulence factors
This is the first report of molecular characterization of S. aureus collected from HD patients in Ha’il region of Saudi Arabia. A high prevalence of MRSA in HD patients as well as high percentage of biofilm production in MRSA isolates were observed in this study. This study emphasizes on the vital role for standardized surveillance along with validated molecular characterization methods to evaluate the incidence of MRSA and accordingly to control its spread among HD patients.
This study was funded by King Abdulaziz City for Science and Technology (KACST), General Directorate of Research Grant 143-34.
The authors declare that there are no conflicts of interest.
- Pollitt, E.J.G., Szkuta, P.T., Burns, N., Foster, S.J. Staphylococcus aureus infection dynamics. PLoSPathog, 2018; 14(6): e1007112.
- Wertheim, H.F., Melles, D.C., Vos, M.C., van Leeuwen, W., van Belkum, A., Verbrugh, H.A., Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis., 2005; 5:751-762.
- Crosby, H.A., Kwieciסski, J., Horswill, A.R. Staphylococcus aureus aggregation and coagulation mechanisms, and their function in host-pathogen interactions. Adv. Appl. Microbiol, 2016; 96: 1-41.
- Rubיn Arturo Silvero Isidre., Acosta, F.R., Vargas, C.R.C., Romero, G.A.V., Perrota, J.F.P., Fretes, R.M.G. Molecular Characterization of Staphylococcus aureus Isolates Obtained from Hemodialyzed Patients at the Hospital de Clםnicas of Paraguay: A pilot study. Int. J Med Stud, 2017; 5(1):14-19.
- Shinefield, H., Black, S., Fattom, A., Horwith, G,.Rasgon, S., Ordonez, J., Yeoh, H., Law, D., Robbins, J.B., Schneerson, R., Muenz, L., Fuller, S., Johnson, J., Fireman, B., Alcorn, H., Naso, R. Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis. N. Engl. J. Med, 2002; 346:491-496.
- Enright, M.C., Day, N.P.J., Davies, C.E., Peacock, S.J., Sprat,t B.G. Multilocus Sequence Typing for Characterization of Methicillin-Resistant and Methicillin-Susceptible Clones of Staphylococcus aureus. J Clin Microbiol, 2000; 38(3):1008-1015.
- Saxena, A.K., Panhotra, B.R. The Prevalence of Nasal Carriage of Staphylo-coccus aureus and Associated Vascular Access-Related Septicemia Among Patients on Hemo-dialysis in Al-Hasa Region of Saudi Arabia. Saudi J. Kidney Dis. Transpl., 2003; 14(1):30-38.
- Sanavi, S., Ghods, A., Afshar, R. Catheter associated infections in hemodialysis patients. Saudi J. Kidney Dis. Transpl., 2007; 18(1):43.
- Churchill, D.N., Taylor, D.W., Cook, R.J., LaPlante, P., Barre, P. Canadian Hemodialysis Morbidity Study. Am J. Kidney Dis., 1992; 19: 214-234.
- del Rio, A., Cervera, C., Moreno, A., Moreillon, P., Mirף, J.M. Patients at risk of complications of Staphylococcus aureus bloodstream infection. Clin. Infect. Dis., 2009; 48: S246-S253.
- Bradley, J.R., Evans, D.B., Calne, R.Y. Long-term survival in haemodialysis patients. Lancet, 1987; 1:295-296.
- Stern, A., Sachdeva, S., Kapoor, R., Singh, J., Sachdeva, S. High blood pressure in dialysis patients: Cause, pathophysiology, influence on morbidity, mortality and management. J. Clin. Diagn. Res., 2014; 8:,ME1-ME4.
- Lowy, F. Staphylococcus aureus infections. N. Engl. J. Med. 1998; 339: 520-532.
- Tong, S.Y., Davis, J.S,, Eichenberger. E., Holland, T.L., Fowler, V.G.Jr.Staphylococcus aureus infections: epidemiology, patho-physiology, clinical manifestations, and management. Clin Microbiol Rev, 2015; 28:603-661.
- Ralph Corey, G. Staphylococcus aureus Bloodstream Infections: Definitions and Treatment.Clinical Infectious Diseases, 2009; 48(4): S254-S259.
- Weems, J.J. Jr. The many faces of Staphylococcus aureus infection: recognizing and managing its life-threatening manifestations.Postgrad Med, 2001; 110: 24-31.
- Imaizumi, T., Hasegawa, T., Nomura, A., Sasaki, S., Nishiwaki, H., Ozeki T., Shimizu, H., Minatoguchi, S., Yamakawa, T., Yazawa, M., Uchida, D., Kawarazaki, H., Miyamoto, M., Suzuki, T.,Koitabashi, K., Furusho, M., Fujita, Y. Association between Staphylococcus aureus bacteremia and hospital mortality in hemodialysis patients with bloodstream infection: a multicenter cohort from Japanese tertiary care centers. Ther.Apher. Dial., 2017; 21:354-360.
- Patel,P.R,,Kallen, A.J., Arduino, M.J. Epidemiology, surveillance, and prevention of bloodstream infections in hemodialysis patients. Am. J. Kidney Dis., 2010; 56:566-577.
- Dialysis in the Kingdom of Saudi Arabia. Saudi J. Kidney Dis. Transpl., 2018; 29(4): 1012-1020.
- Heilmann, C., Gerke, C., Perdreau-Remington., Go¨tz, F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun, 1996; 64:277-282.
- Al-Abbas, M.A. Antimicrobial susceptibility of Enterococcus faecalisand a novel Planomicrobium isolate of bacterimia. International Journal of Medicine and Medical Sciences, 2012; 4(2): 19-27.
- Tokajian, S., Eisen, J.A., Jospin, G., Farra, A., Coil, D.A. Whole genome sequencing of extended-spectrum b-lactamase producing Klebsiella pneumoniae isolated from a patient in Lebanon. Front Cell Infect Microbiol, 2015; 5: 32.
- Dopirak, M., Hill, C., Oleksiw, M.,Dumigan, D., Arvai, J., English, E., Carusillo, E., Malo-Schlegel, S., Richo, J., Traficanti, K., Welch, B., Cooper, B. Surveillance of hemodialysis-associated primary bloodstream infections: the experience of ten hospital-based centers. Infect Control Hosp Epidemiol, 2002; 23:721-724.
- Klevens, R.M, Edwards, J.R., Andrus, M.L., Peterson, K.D., Dudeck, M.A., Horan, T.C., NHSN Participants in Outpatient Dialysis Surveillance. Dialysis surveillance report: National Healthcare Safety Network (NHSN) – Data summary for 2006. Semin Dial, 2008; 21:24-8.
- Archer, N.K., Archer, N.K., Mazaitis, M.J., Costerton, J.W., Leid, J.G., Powers, M.E., Shirtliff, M.E. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence, 2011; 2:445-459.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.