Methicillin-resistant Staphylococcus aureus (MRSA) has emerged as a persistent pathogen in the seafood industry, raising concerns for food safety. Identifying the origin and transmission of MRSA is highly critical to control its spread, thereby acts as a potential tool for virtual networking of MRSA transmission in seafood. The study was conducted in the seafood commodities across two fish landing centres and three adjoining retail fish markets of Kottayam district, Kerala. Seventy-four samples were collected by a set sampling plan and the samples were repeated for validating transmission tracking. The Staphylococcal protein A (spa) typing, clumping factor B, capsular polysaccharide and accessory gene regulator typing was accomplished on MRSA isolates to identify the type of strain present in the sample and their probable dissemination between the fish landing centres to retail fish markets. The molecular typing tools categorized the MRSA isolates into CP5-clfb-(t334), CP8-clfb-(t304, t3841, t304 and t127). Discriminatory power (D) was highest for spa typing than others tools. The study confirms the existence of MRSA in seafood and notices that MRSA are carried through seafood from landing centres to their adjoining retail markets. MRSA, owing to its recently established persistence in the seafood and spa typing tool due to its cost-effectiveness can be used in combination for the virtual networking of the transmission of seafood. Prior to this, the diversity of each location needs to be mapped.
MRSA, Seafood, Virtual Networking, Spa Typing
Microflora of the fish in retail outlets are a reflection of general contamination in the aquatic environment as well as post-harvest handling contamination.1 Harvesting, storage, distribution, processing, and preservation of seafood are the major handling practice responsible for the transmission of microbes from fish to human and vice versa.2-6 The possibility of transmission of MRSA between humans and food has been described.7,8 Similarly, augmented reports have established MRSA in various food-producing industries. Since 1961, MRSA has been a huge problem in the health care settings owing to it gaining resistance to the most potent antibiotics.9 In seafood, MRSA enters by post-harvest contamination and flourishes as a persistent pathogen. However, genetic level comparison of the MRSA isolates revealed that uniqueness existed between the hospital, community, livestock, and environmental sectors.6,10-12 Hence, specific strains of MRSA have become persistent in a particular population or environment.5 In contrast to the S. aureus which is already an established bacterial population in the environment, MRSA is recently evolving as a persistent population in the food sector. Hence, there is a possibility of using this microbial population as a virtual networking tool, and thereby link the MRSA transmission in the seafood supply chain as well as assigning geographical identity. Even though bacterial source tracking studies are implemented for various indicator bacteria and gram-negative bacteria, benefit of using MRSA as source tracking tool was not performed.13
Based on MRSA strain, its origin and dissemination can be investigated through local epidemiological approach. Generally, micro-variation analyses are being used for local epidemiological studies in a particular environment/population and in outbreak investigations.14 In the case of MRSA, genetic typing tools available for local and global epidemiology are staphylococcal protein A (spa) typing, clumping factor B (Clfb) typing, capsular protein (CP) typing. accessory gene regulator (agr) typing pulsed-field gel electrophoresis (PFGE) and Multi-locus sequence typing (MLST).5,15-18 However, the tools become more popular because of their user friendliness and economic viability.19 The applicability of these tools for microbial source tracking of contamination was proven in hospital sector and in seafood.5,20,21
Hence, in the present investigation, spa typing, virulence factor typing (clfb + CP) and agr typing were used as characterization tools as well as for tracking the contamination. The key idea behind this study is to validate the tools for source tracking the MRSAs in the seafood with earlier documented different sets of landing centre and retail markets samples5 which can be used as the virtual networking tool for a better predicting the origin of the food from geographically distinct locations based on the strain identification.
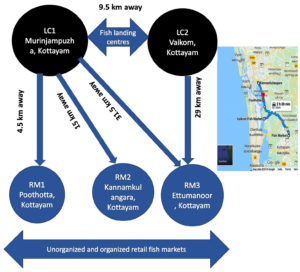
The sampling plan consisted of two traditional fish landing centres and three retail fish markets which include unorganized roadside as well as organized markets of Kottayam District, Kerala, India,5 not covered in the earlier study (Figure 1). The distances between fish landing centre and retail fish markets ranged from 4.5 km to 29 km. Sampling was carried out early in the morning at fish landing centre followed by fish markets at random. The study was carried out in four batches with an average of 7 samples of water and fishes from fish landing centre and 7 samples from the retail market. The sampling strategy, based on the availability, envisages to include one finfish, shellfish, mollusc, water and ice used at every landing centre and retail fish market. Totally, 72 samples were collected during the entire study, which includes finfish, crustaceans, molluscs, water and ice (Table 1). Samples were collected in sterile bags (labelled landing centre and fish market wise) and brought to the laboratory in the insulated icebox and processed within four hours of sample collection.
Figure 1. Study area and distance between fish landing centres and retail fish markets, Kottayam, Kerala, India
LC1: Landing centre 1- Murinjampuzha; LC2: Landing centre 2- Vaikom; RM1: Retail Market 1- Unorganized Poothotta fish market; RM2: Kannamkulangara unorganized fish market, RM3: Organized retail fish market Ettumanoor. The distance between two landing centre is 9.5 km; The minimum distance between landing centre to retail markets is 4.5 km and a maximum of 29 km (Source: Google Maps)
Table (1):
Details of samples collected in Batch I to IV between fish Landing centres and retail markets of Kottayam District, Kerala
| Batch | Name of landing centre, retail fish market and fish species |
|---|---|
| I | Murinjampuzha (Landing Centre 1; LC1): |
| Samples – Sardinella longiceps, Dried Salted Cod, Etroplus suratensis, Scylla serrata, Shrimp Fenneropenaeus indicus, Water and ice | |
| Poothotta (Roadside Market 1; RM1): | |
| Samples – Sardinella longiceps and ice | |
| II | Vaikom (Landing Centre 2; LC 2): |
| Samples – Fenneropenaeus indicus, Dried Salted Mackerel, Chanos chanos and Mystus oculatus sp. | |
| Kannamkulangara, (Retail market 2; RM2): | |
| Samples –Villorita cyprinoides, Penaeus monodon, Rastrelliger kanagurta, Bramidae sp. and ice | |
| Murinjampuzha (Landing Centre 1; LC1): | |
| Samples – Villorita cyprinoides, Sardinella longiceps, Mixed Dried Salted Fish, Sardinella longiceps, Etroplus suratensis, Penaeidae, Oreochromis niloticus, Sphyraena barracuda, Scylla serrata, Dendrobranchiata sp., Ice and Water | |
| III | Vaikom (Landing Centre 2; LC2) |
| Samples – Villorita cyprinoides, Dendrobranchiata, Stolephorus indicus, Mixed Salted Dried Fish, Siluriformes sp., Sardinella longiceps, Etroplus suratensis, Chanos chanos, Scylla serrata, Water, Ice, and Ice with Fish | |
| Ettumanoor (Retail Market 3; RM3): | |
| Samples – Sardinella longiceps, Villorita cyprinoides, Gadus morhua and Dendrobranchiata | |
| IV | Murinjampuzha (Landing Centre 1; LC1): |
| Samples – Lutjanus campechanus, Villorita cyprinoides, Mixed Fish, Brachyura sp., Engraulidae sp., Etroplus suratensis, Sardinella longiceps and Water | |
| Stolephorus indicus, Dried Prawns, Mystus oculatus, Scylla serrata, Fenneropenaeus indicus, Water and Ice | |
| Ettumanoor (Retail Market 3; RM3): | |
| Samples – Villorita cyprinoides, Stolephorus indicus, Penaeus monodon, Sardinella longiceps, Etroplus maculatus, Etroplus suratensis, Selar crumenophthalmus, Lactarius lactarius and Ice |
MRSA was isolated and identified from the seafood samples as per the Bacteriological Analytical Manual with modifications.3,22 Initially, all the isolates were confirmed conventionally by Oxacillin agar dilution method23 then molecularly by multiplex PCR (mPCR) targeted genes specific for S. aureus (nuc), MRSA (mec) and 16S rDNA for Staphylococci.24 Further, the molecular tools viz. spa typing, virulence Factor typing (clfb + CP) and agr typing were used to characterize the strains of MRSA isolated. In all the PCR assays, S. aureus ATCC 43300 was used as MRSA positive control, S. aureus ATCC 29213 was used as negative control for methicillin resistance, and no template control was used for reaction negative control.
Using spa-1113f and spa-1514r primers, the variable region within the conserved region of the spa gene was amplified for detecting the presence of spa gene of variable sizes.25 The amplified region was eluted and outsourced for sanger sequencing (Genespec Pvt. Ltd., India) and the spa types were deduced from ridom server as well as Fortinbras software available in the public domain and same was used to characterize the isolates from all the sectors of the source without any modification.4
A virulence factor typing was carried out with the combination of clfb and cap to characterize the MRSA. The pathogenesis is significantly influenced by capsular polysaccharide (CP), which is produced by S. aureus. More than 80% of clinical isolates produce either CP5 or CP8 capsular polysaccharides.26,27 DNA templates (3 µl) were added to 22 µl of PCR mix containing 1x conc. of Master Mix (Thermo Scientific) and 0.4 µM of each primer. PCR conditions included an initial denaturation stage at 94 °C for 5 minutes, 35 cycles of 94 °C for 30 seconds, {49.0 °C for 30 seconds (cap5 gene)/46.0 for 30 seconds (cap8 gene)}, and 72 °C for 1 minute, and a final extension step at 72 °C for 7 minutes comprised the amplification cycle.
PCR was performed for agr typing for all isolates using primers agr I, agr II, agr III, and agr IV in a 25 µl reaction volume containing 0.4 mM dNTP’s, 0.6 µM concentration of each primer (agr pan F, agr pan I, agr pan II, agr pan III and agr pan IV, respectively), 2.5 µl of 10X buffer, 3 mM MgCl2, 1 U/µl of EX Taq DNA polymerase (Takara) and template DNA of 3 µl. PCR conditions included an initial denaturation step at 94 °C for 5 minutes, 35 cycles of 94 °C for 30 seconds, 55 °C for 30 seconds, and 72 °C for 1 minute, and a final extension step at 72 °C for 10 minutes.28,29
The discriminatory power of each method in delineating seafood environment MRSA were calculated using the method described.30 Briefly calculating how many types of MRSA can be categorized from each typing method with the total number of strains used for characterization was calculated based on the formula described below,
D = 1 – 1/(N(N-1) ∑sj=1 xj (xj -1)
Where D = Index of Discriminatory Power, N = Number of unrelated strains tested, S = Number of different types and xj = Number of strains belonging to the jth type”.
To check for the existence of MRSA isolates, 74 samples from different fish landing centres and retail fish marketplaces were used in this investigation. The multiplex PCR were used for the molecular confirmation of MRSA by targeting 16S rRNA of 756 bp (Genus specific for Staphylococcus), nuc of 279 bp (Species specific for S. aureus), and mecA of 310 bp (a gene for methicillin resistance).24 The isolates which showed all three genes were confirmed as MRSA.
A total of 12.16% of samples showed positive result for MRSA. This included 6.75% from the fish landing centres (LC) and 5.40% from the retail fish markets (RM). Among that 1.35% of samples were isolated from LC2, 1.35% from RM1, 5.40% from LC1 and 4.05% from RM3. LC2 and RM1 showed the lowest number of MRSA positive samples. The study indicates that MRSA is becoming persistent in the seafood niche and understanding its diversity on a larger scale might be potentially useful for tracking studies (Table 2). Before the strains of MRSA get established as a persistent population in the seafood sector, the strain assignment would provide more scope for the networking of food transmission.
The presence of clfB and cap5 or cap8 indicates the potential colonization of these MRSA’s in the vestibulum nasi of human beings, adhesion to squamous epithelium, adhesion to a or b chains of human fibrinogen and also aids the organism to survive from the uptake and killing by host neutrophils.31,32 Based on the presence of virulence factors (cap and clfb), the MRSA were categorized to cap5 & clfb and cap8 & clfb (Table 2). Accessory gene regulatory (agr) genes are the important regulator for controlling the expression of virulence factors in S. aureus and based on the sequence variations in the genes, agr are divided into agrC (autoinducing peptide) and agrD (cyclic AIP), and further categorized into four groups (agr I, agr II, agr III, and agr IV). Based on the agr typing, the MRSAs isolated in this study belonged to only agrI (Table 2).
Table (2):
Spa Types identified in fish landing centre and retail markets
No. |
Sampling site |
Samples |
Spa types |
Virulence types (Cap + clfb type) |
agr type |
|---|---|---|---|---|---|
1 |
Murinjampuzha Landing centre (LC1) |
Ice |
t3841 |
Cap8 + clfb |
agr I |
2 |
Murinjampuzha Landing centre (LC1) |
Clam |
t334 |
Cap5 + clfb |
agr I |
3 |
Murinjampuzha Landing centre (LC1) |
Pearl Spot |
t304 & t311 |
Cap8 + clfb & Cap5 + clfb |
agr I & – |
4 |
Murinjampuzha Landing centre (LC1) |
Barracuda |
t334 |
Cap5 + clfb |
agr I |
5 |
Vaikam Landing centre (LC2) |
Indian Prawns |
t3841 & t304 |
Cap8 + clfb & Cap8 + clfb |
agr I & agr I |
6 |
Poothotta Retail market (RM1) |
Ice |
t304 |
Cap8 + clfb |
agr I |
7 |
Ettumanoor Retail market (RM3) |
Ice |
t304 |
Cap8 + clfb |
agr I |
8 |
Ettumanoor Retail market (RM3) |
Clam |
t3841 & t304 |
Cap8 + clfb & Cap8 + clfb |
agr I & agr I |
9 |
Ettumanoor Retail market (RM3) |
False Trevally |
t334 & t127 |
Cap5 + clfb & Cap8 + clfb |
agr I & – |
The samples of fish/clam or ice detected with MRSA and their spa types; Combination virulence and agr profiles identified in two landing centres and two retail markets
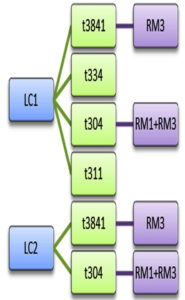
Sanger sequence analysis showed 5 different spa types among the 36 isolates of MRSA viz., spa types t3841, t334, t127, t304 and t311. According to the spa typing results, t3841, t334 and t304 were the predominant spa types. The strains t3841 and t304 were found in both LC1 and LC2 and in RM1 and RM3 samples. Additionally, LC1 harboured t334 and t311 spa types and RM3 harboured t127 and t334 spa types. Pearl spot from LC1, Indian prawns from LC2 and False Trevally from RM3 carried 2 types of MRSA clones in a single seafood sample (Figure 2). This shows the diversity of the pathogenic organism in a single sample itself and t304 was present in both the landing centres and retail markets. This implicates the dissemination of microorganisms from the landing centre to the adjoining retail markets. t311 isolated from pearl spot collected from LC1 showed no evidence of dissemination; similarly, t127 isolated from False Trevally also showed no evidence of the origin of the source.
Figure 2. Dissemination Pattern of MRSA clones between fish landing centres and retail fish markets. The spa types t3841, t334 and t304 detected at landing centre were carried between landing centre and retail markets. t304 was detected both in Landing centre 1 and 2 as well as retail market 1 and 3. t3841 was detected in both landing centres and in retail market 3
Clfb typing along with spa typing have the capability of discriminating isolates within clonal groups. 13, 33 The most predominant capsular polysaccharide serotypes are CP5 and CP8 34 and in the study both of these types were identified as CP5 (t334) at 25% and CP8 (t304, t3841, t304 and t127) at 75% incidence.
Among MRSA isolates, spa typing had the highest discriminating power (D = 0.758), followed by agr typing (D = 0.4) and cap typing (D = 0.3555). A D value of 0.0 in clfb typing implies that each and every member of a strain population belonged to the same type (Table 3). Poor hygiene and inadequate sanitization during the processing leads to MRSA contamination.35 in ready to eat, ready to cook and fresh seafood.3-6,36,37 MRSA is evolving in the seafood environment and it is becoming a persistent population.
Table (3):
Discriminatory Index of Each Typing Methods
Typing Method |
Discriminatory Indexes, D |
Cost (App USD) |
Time (hrs) |
|---|---|---|---|
spa typing |
0.7508 |
15 |
1.7083 |
agr typing |
0.4 |
5 |
1.4160 |
clfb typing |
0.0 |
5 |
1.3666 |
cap typing |
0.3555 |
5 |
1.3666 |
One of the best methods for controlling and preventing the spread is molecular typing. The ideal molecular typing test for epidemiological research should be simple to use, affordable, highly reproducible, quick, and simple to interpret.38 The present study represents the simultaneous tool comparison for molecular source tracking of foodborne antibiotic-resistant pathogen (MRSA) between landing centres and retail markets, and this provides an insight into the diversity of genotypes of MRSA. The various MRSA typing tools such as PFGE, MLST, SCCmec typing are expensive, time-consuming and may be difficult to interpret.25 But the spa typing of MRSA allows speed, costs low and provides comparability of data which are the essential factors for the source tracking of MRSA in seafood chain. clfB, CP (5&8) and agr typing methods could explain the clinical characteristics of the MRSA isolates.39 The combined results of spa typing and clfB typing are highly stable and has high discriminatory power comparatively to PFGE and whole-genome microarray analysis.14 The CP typing plays a major role in the immunotherapy against pathogen and the prevalence of capsular types 5 and 8 varied between the isolates.40 There is an obvious correlation between capsular type and genotype, according to the findings of the CP and spa typing tests. Some researchers have found that the expression of the agr gene is differentially regulated in various S. aureus groups and is involved in the regulation of the production of bacterial virulence factors. Agr I type isolates make up 77.77% of all isolates in this investigation. The key idea behind this study is to assess the seafood contamination by setting MRSA as candidate organism and spa typing as a molecular marker to assess the diversity and also transmission between the landing centre and its adjoining retail markets. Once the diversity of all the fish landing centres and retail fish markets are established for MRSA, it can be virtually networked for understanding the movement of the food between geographical locations using MRSA and spa typing tool. Our previous study was carried out as source tracking the contamination of MRSA between landing centre and retail market of Kottayam and Ernakulam districts with the help of MLST and spa typing tool, however in the present study only Kottayam district landing centres and retail markets were studied with spa typing and other PCR based virulence markers tool.4
The bacterial persistence was initially observed in Staphylococci with penicillin exposure in early 1942 and the surviving bacterium became the persistent population over the years.41 So, the persistent population can rapidly adapt to the antibiotic stress by triggering gene responses linked with various stressors.42 The increased use of antibiotics eventually leads to the sudden spread of MRSA and its possible transmission is greatly dependent on its persistence in the environment.43 The rapid increase of MRSA clones in the aquatic environment could be an indication that MRSA clones are getting established as persistent and therefore can be used as markers. Only a comprehensive investigation to identify the source and dissemination pattern of contamination could assure seafood safety.
In this persistent MRSA, diversity is documented completely between various landing centres and retails markets in the country. In this virtual networking process, the genetic identity of this persistent and evolving MRSA needs to be documented in over 1500 fish landing centres across India along with their adjoining retail fish markets so that it will provide valuable information. This study also compares the tools to study the molecular epidemiology of MRSA isolates that can be used as a molecular source tracking indicator. This study indicates 12.16% of samples contain MRSA isolates and can be used as a benchmark for future studies. In the extended period of time, the same pathogen can be used as an indicator pathogen for molecular source tracking. Individual molecular markers could not provide 100% source identification due to a lack of sensitivity and specificity.44 Although MRSA does not constitute the normal flora of fresh seafood, it could become a part of persistent populations.45 By considering the current population of MRSA as a molecular marker, the emergence and transformation of MRSA as a persistent population can be identified. The use of spa typing alone could provide evidence of the prevalence, diversity, and dissemination of MRSA clones between the landing centres and retail markets. Very few studies have focused on MRSA in seafood and many of them concentrated only on the prevalence and diversity of MRSA.46,47 Other studies mainly explain the antibiotic resistance, epidemiological types and virulence profile of MRSA isolated from marine fish.48 Source tracking study explicitly used on animal samples for phylogenetic tracking of LA-MRSA.49 Hence, in this study, we have used this organism as an indicator organism and we compared molecular tools for characterizing them to strain type, and also tried to document these types of MRSA available in the sector. This molecular tool not only documents diversity but also confirms the movement of the seafood from one landing centre to retail markets. This would make this study unique and relevant from various existing studies. This is the first study to conceptualize the food transmission with the bacterial source tracking tool being MRSA and its molecular typing tools.
The study was conducted for 74 samples representing two fish landing centres and three retail fish markets, in order to better understand the local epidemiological picture. However, understanding the broader epidemiology, i.e. between the districts and between the states, there shall be a network project encompassing wider geographical regions covering all the major fish landing centres and retail fish markets representing the major points of seafood harvest and distribution. Covering wider geographical region with representative landing centres and retail markets will validate this tool’s suitability in the virtual networking of seafood transmission. Also, the study may be explored for the use of several phenotypic fingerprinting tools like antimicrobial resistance to supplement the source tracking protocol.
The study concludes that there is a prevalence of 12.16% MRSA in samples screened in this study in different landing centres and retail markets. The diversity of MRSA clones showed the complexity in the epidemiology of the organism. The result reveals the details regarding the dissemination of MRSA between the landing centres and the retail markets. Hence, it is extremely important to perform virtual networking of transmission of MRSA in the seafood chain by keeping the population as a standard and cautions fish handlers to follow stringent sanitary procedures during harvesting, handling, and storing of fish.
ACKNOWLEDGMENTS
The authors are thankful to the Director, ICAR-CIFT, Cochin, for providing support for the conduct of the research and are also thankful to ICAR for funding the research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AT, MV, MRB, TCJ and MPM conceptualized the study. AT, MV, and VS, ENS performed sample collection. AT, RK and SVN conducted the experiments. AT and MV carried out the sequencing analysis. AT and MV wrote the manuscript. AT, MV, VS and ENS edited the manuscript. AT, MV, MRB, TCJ and MPM reviewed and approved the final manuscript for publication.
FUNDING
The research work is carried out under ICAR-CIFT funded Institutional grant.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Pal M, Ketema A, Anberber M, Mulu S, Dutta Y. Microbial quality of Fish and Fish Products. Beverage & Food World. 2016;43(2):1-4.
- Gates, K. W. (2010). Fishery Products—Quality, Safety and Authenticity: Edited by H. Rehbein and J. Oehlenschläger. Chichester, West Sussex, UK. Wiley-Blackwell, John Wiley and Sons, Ltd., Publication, 2009:477. Journal of Aquatic Food Product Technology, 19(3-4):318-325.
Crossref - Murugadas V, Joseph TC, Reshmi K, Lalitha KV. Prevalence of methicillin resistant Staphylococcus aureus in selected seafood markets and aquaculture farms in Kerala, south-west coast of India. Indian J Fish. 2016;63(4):150-153.
Crossref - Murugadas V, Toms CJ, Reethu SA, Lalitha KV. Multilocus sequence typing and staphylococcal protein a typing revealed novel and diverse clones of methicillin-resistant Staphylococcus aureus in seafood and the aquatic environment. J Food Prot. 2017;80(3):476-481.
Crossref - Murugadas V, Joseph TC, Lalitha KV. Tracing contamination of Methicillin-resistant Staphylococcus aureus (MRSA) into seafood marketing chain by staphylococcal protein A typing. Food Control. 2017b;78:43-47.
Crossref - Vaiyapuri M, Joseph TC, Rao BM, Lalitha KV, Prasad MM. Methicillin-Resistant Staphylococcus aureus in Seafood: Prevalence, Laboratory Detection, Clonal Nature, and Control in Seafood Chain. J Food Sci. 2019;84(12):3341-3351.
Crossref - De Sousa MA, Sanches IS, Van Belkum A, Van Leeuwen W, Verbrugh H, De Lencastre H. Characterization of methicillin-resistant Staphylococcus aureus isolates from Portuguese hospitals by multiple genotyping methods. Microb Drug Resist. 1996;2(3):331-341.
Crossref - Jones TF, Kellum ME, Porter SS, Bell M, Schaffner W. An outbreak of community-acquired foodborne illness caused by methicillin-resistant Staphylococcus aureus. Emerg Infect Dis. 2002;8(1):82-84.
Crossref - Archer GL. Staphylococcus aureus: A Well-Armed Pathogen. Clin Infect Dis. 1998;26(5):1179-1181.
Crossref - Montgomery CP, Boyle-Vavra S, Daum RS. Importance of the global regulators agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS ONE. 2010;5(12):1-9.
Crossref - McClure JAM, Lakhundi S, Kashif A, Conly JM, Zhang K. Genomic comparison of highly virulent, moderately virulent, and avirulent strains from a genetically closely-related MRSA ST239 sub-lineage provides insights into pathogenesis. Front Microbiol. 2018;9:1-16.
Crossref - Turner NA, Sharma-Kuinkel BK, Maskarinec SA, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. 2019;17(4):203-218.
Crossref - Foley SL, Lynne AM, Nayak R. Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect Genet Evol. 2009;9(4):430-440.
Crossref - Koreen L, Ramaswamy SV, Naidich S, Koreen IV, Graff GR, Graviss EA, Kreiswirth BN. Comparative Sequencing of the Serine-Aspartate Repeat-Encoding Region of the Clumping Factor B Gene (clfB) for Resolution within Clonal Groups of Staphylococcus aureus. J Clin Microbiol. 2005;43(8):3985-3994.
Crossref - Laabei M, Recker M, Rudkin JK, et al. Predicting the virulence of MRSA from its genome sequence. Genome Res. 2014;24(5):839-849.
Crossref - Verhegghe M, Pletinckx LJ, Crombe F, et al. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 in pig farms and multispecies farms. Zoonoses Public Health. 2013;60(5):366-374.
Crossref - Liu Q, Han L, Li B, Sun J, Ni Y. Virulence characteristic and MLST-agr genetic background of high-level mupirocin-resistant, MRSA isolates from Shanghai and Wenzhou, China. PLoS ONE. 2012;7(5):1-8.
Crossref - Krzyszton-Russjan J, Tambic-Andrasevic A, Bukovski S, Sabat A, Hryniewicz W. First community-acquired methicillin-resistant Staphylococcus aureus (MRSA) strains in Croatia. Clin Microbiol Infect. 2006;12(7):697-698.
Crossref - Ostojic M, Hukic M. Genotypic and phenotypic characteristics of methicillin-resistant Staphylococcus aureus (MRSA) strains, isolated on three different geography locations. Bosn J Basic Med Sci. 2015;15(3):48-56.
Crossref - Khandavilli S, Wilson P, Cookson B, Cepeda J, Bellingan G, Brown J. Utility of spa typing for investigating the local epidemiology of MRSA on a UK intensive care ward. J Hosp Infect. 2009;71(1):29-35.
Crossref - Ho J, Boost MV, O’Donoghue MM. Tracking sources of Staphylococcus aureus hand contamination in food handlers by spa typing. Am J Infect Control. 2015;43(7):759-761.
Crossref - Tallent S, Hait J, Bennett RW (ret.), Lancette GA, Bacteriological Analytical Manual Chapter 12: Staphylococcus aureus, AOAC International, Gaithersburg, Md., 1998. Acessed on December, 12, 2024
- Ghanwate N, Thakare P, Bhise PR, Gawande S. Colorimetric method for rapid detection of Oxacillin resistance in Staphylococcus aureus and its comparison with PCR for mec A gene. Sci Rep. 2016;6:1-5.
Crossref - Zhang K, Sparling J, Chow BL, et al. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J Clin Microbiol. 2004;42(11):4947- 4955.
Crossref - Hashemizadeh Z, Bazargani A, Kalantar-Neyestanaki D, Mohebi S, Hadi N. Determining spa-type of methicillin-resistant Staphylococcus aureus (MRSA) via high-resolution melting (HRM) analysis, Shiraz, Iran. BMC Res Notes. 2020;13(1):1-4.
Crossref - O’Riordan K, Lee JC. Staphylococcus aureus Capsular Polysaccharides. Clin Microbiol Rev. 2004;17(1):218-234.
Crossref - Ote I, Taminiau B, Duprez JN, Dizier I, Mainil JG. Genotypic characterization by polymerase chain reaction of Staphylococcus aureus isolates associated with bovine mastitis. Vet Microbiol. 2011;153(3-4):285-292.
Crossref - Gilot P, Lina G, Cochard T, Poutrel B. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J Clin Microbiol. 2002;40(11):4060-4067.
Crossref - Xie Y, He Y, Gehring A, et al. Genotypes and toxin gene profiles of Staphylococcus aureus clinical isolates from China. PloS one. 2011; 6(12):e28276.
Crossref - Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J Clin Microbiol. 1988;26(11):2465-2466.
Crossref - Eldhin DN, Perkins S, Francois P, Vaudaux P, Hook M, Foster TJ. Clumping factor B (ClfB):a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol Microbiol. 1998;30(2):245-257.
Crossref - Wertheim HFL, Walsh E, Choudhurry R, et al. Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med. 2008;5(1):0104-0112.
Crossref - Dai Y, Liu J, Guo W, et al. Decreasing methicillin-resistant Staphylococcus aureus (MRSA) infections is attributable to the disappearance of predominant MRSA ST239 clones, Shanghai, 2008-2017. Emerg Microb Infect. 2019;8(1):471-478.
Crossref - Liu B, Park S, Thompson CD, Li X, Lee JC. Antibodies to Staphylococcus aureus capsular polysaccharides 5 and 8 perform similarly in vitro but are functionally distinct in vivo. Virulence. 2017;8(6):859-874.
Crossref - Cho JIl, Joo IS, Choi JH, et al. Distribution of Methicillin-resistant Staphylococcus aureus (MRSA) in RAW meat and fish samples in Korea. Food Sci Biotechnol. 2014;23(3):999-1003.
Crossref - Abrahim A, Sergelidis D, Kirkoudis I, et al. Isolation and antimicrobial resistance of Staphylococcus spp. in freshwater fish and greek marketplaces. J Aquat Food Prod Technol. 2010;19(2):93-102.
Crossref - Visnuvinayagam S, Joseph TC, Murugadas V, Chakrabarti R, Lalitha KV. Status on methicillin resistant and multiple drug resistant Staphylococcus aureus in fishes of Cochin and Mumbai coast, India. J Environ Biol. 2015;36(3):571-575.
- Fasihi Y, Fooladi S, Mohammadi MA, Emaneini M. The spa typing of methicillin-resistant Staphylococcus aureus isolates by High Resolution Melting (HRM) analysis. J Med Microbiol. 2017;66(9):1335-1337.
Crossref - Ruppitsch W, Indra A, Stoger A, et al. Classifying spa Types in Complexes Improves Interpretation of Typing Results for Methicillin-Resistant Staphylococcus aureus. J Clin Microbiol. 2006;44(7):2442-2448.
Crossref - Melles DC, Taylor KL, Fattom AI, van Belkum A. Serotyping of Dutch Staphylococcus aureus strains from carriage and infection. FEMS Immunol Med Microbiol. 2008;52(2):287-922.
Crossref - Hobby GL, Meyer K, Chaffee E. Observations on the mechanism of action of penicillin. Proceedings Society Exp Biol and Med. 1942;50(2): 281-285.
Crossref - Gefen O, Balaban NQ. The importance of being persistent: Heterogeneity of bacterial populations under antibiotic stress: Review article. FEMS Microbiol Rev. 2009;33(4):704-717.
Crossref - Plano LR, Garza AC, Shibata T, et al. Shedding of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus from adult and pediatric bathers in marine waters. BMC Microbiol. 2011;11:5.
Crossref - Balleste E, Bonjoch X, Belanche LA, Blanch AR. Molecular indicators used in the development of predictive models for microbial source tracking. Appl Environ Microbiol. 2010;76(6):1789-1795.
Crossref - Raymond A, Ramachandran A. Bacterial Pathogens in Seafood-Indian Scenario. Fishery Technology. 2019;56:1-22.
- Fri J, Ndip RN, Njom HA, Clarke AM. First report of methicillin-resistant Staphylococcus aureus in tank cultured dusky kob (Argyrosomus japonicus):and evaluation of three phenotypic methods in the detection of MRSA. Journal of Food Safety. 2018;38(1):1-6.
Crossref - Ayulo AMR, Machado RA, Scussel VM. Enterotoxigenic Escherichia coli and Staphylococcus aureus in fish and seafood from the southern region of Brazil. Int J Food Microbiol. 1994;24(1-2):171-178.
Crossref - Fri J, Njom HA, Ateba CN, Ndip RN. Antibiotic Resistance and Virulence Gene Characteristics of Methicillin-Resistant Staphylococcus aureus (MRSA) Isolated from Healthy Edible Marine Fish. Int J Microbiol. 2020;2020(1):9803903.
Crossref - Lienen T, Schnitt A, Cuny C, Maurischat S, Tenhagen BA. Phylogenetic tracking of LA-MRSA ST398 intra-farm transmission among animals, humans and the environment on German dairy farms. Microorganisms. 2021;9(6):1119.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.