ISSN: 0973-7510
E-ISSN: 2581-690X
Aromatic amines are potential mutagens and carcinogens, so biodegradation of them may protect our environment by safely getting rid of these compounds. In the present study, 5 aniline degrading isolates were recovered from 30 samples of sewage and agricultural wastewater, The five isolates were identified biochemically and by 16S rRNA gene sequence analysis as Achromobacter sp. (2 isolates), Pseudomonas sp. (2 isolates) and Enterobacter sp. (1 isolate). Biodegradation of aniline was measured by HPLC, and within 10 days, aniline degradation percentage was variable ranging from 27% to 61% of the initial aniline concentration with the formation of catechol as the byproduct which was further biodegraded to cis, cis muconic acid confirming that the isolate was following the ortho-cleavage pathway. Additionally, the ability of these isolates to degrade aniline was confirmed by measuring the optical density (OD) of the growth of the five isolates in the presence of aniline as sole carbon source using the microtiter plate method at 620 nm. The genomic DNA and plasmid of the isolates were then extracted. Screening for selected genes, known to encode aniline depredating enzymes; cat1,2, cat2,3, tadR, and tdnQ using PCR was done. It revealed the presence of cat1,2, tadR, and tdnQ in the plasmid preparation. Significance, and Impact of study: It was concluded that aniline is degraded by the five isolates through a pathway involving enzymes performing ortho-cleavage of the benzene ring and these isolates could have a potential role for use in bioremediation of aniline contaminated environments.
Aniline, Biodegradation, Cat 1,2, Cat 2,3, tadR, tdnQ
Environmental pollution resulting from the incontinent synthesis of xenobiotic compounds has become a dangerous threat to the biosphere (Wang et al., 2007). The second most common class of organic compounds in the biosphere, after carbohydrates are aromatic compounds (Bull et al., 2011). Industrial sectors such as oil refining, dyes, cosmetics, medicines, rubber, agrochemicals, explosives are considered the primary sources of aromatic amines (Palmiotto et al., 2001).
Aromatic amines can be classified into three types: monocyclic, polycyclic and heterocyclic (DeBruin and Josephy, 2002; Stellman, 1998). Monocyclic aromatic amines bring hazards to human health and aniline is one of them.
Aniline, the parent molecule of aromatic amines with C6H5NH2 chemical formula is amongst the most critical substances in chemistry (Sihtmae et al., 2010). The pharmaceutical and industrially manufactured azo-dye are the primary sources for aniline discharge to the environment (Essington, 1994; Rai et al., 2005). Due to the expanded use of aniline in the industry, it released into the soil and water bodies (Zeyer et al., 1985). The high toxicity of aniline and its derivatives is reported to be a significant cause of cancer (Bomhard and Herbold, 2005; Markowitz and Levin, 2004).
One of the strategies of aniline detoxification is aniline biodegradation by soil microorganisms (Pluvinage et al., 2007; Suzuki et al., 2007). Many bacteria able to degrade aniline were isolated from agricultural soils and industrial areas (Liang et al., 2005; Vangnai and Petchkroh, 2007); aerobic and anaerobic degradation pathways of aniline were also investigated (Boon et al., 2001; Carmona et al., 2009; Liang et al., 2005).
The first step in the aerobic degradation pathway is oxidative deamination with the production of catechol, with further degradation to cis, cis-muconic acid by the catechol 1,2-dioxygenase (ortho-cleavage pathway) (Aoki et al., 1984; Loidl et al., 1990; Murakami et al., 2003) or to a yellow compound, 2-hydroxymuconic semialdehyde by catechol 2,3-dioxygenase (the meta-cleavage pathway)(Fujii et al., 1997; Takeo et al., 1998a; Takeo et al., 1998b).
The bacterial species of Pseudomonas (Kahng et al., 2000a; Tanaka et al., 2009), Comamonas (Boon et al., 2000), Rhodococcus (Zhuang et al., 2007), Acinetobacter (Wyndham, 1986), Frateuria (Murakami et al., 2003), Moraxella (Zeyer et al., 1985), Delftia (Kahng et al., 2000a; Zhang et al., 2008) and Nocardia (Wang et al., 2006) have been shown to be efficient degraders of aniline and its derivatives (Liu et al., 2002). These bacteria showed the ability to efficiently biodegrade aniline under neutral and moderate conditions (Li et al., 2010) and up till now, aerobic biological treatment was reported to remove aniline efficiently (Chengbin et al., 2009; Takeo et al., 1998b; Zhang et al., 2008).
The first step in aniline conversion to catechol is a multistep reaction catalyzed by three enzymes, a glutamine synthetase (GS)-like enzyme, aniline dioxygenases and glutamine amidotransferase like enzyme (Liang et al., 2005; Murakami et al., 1998). Firstly, GS like enzyme catalyzed ATP-dependent ligation of L-glutamate to aniline to form gamma-glutamylanilide (Takeo et al., 2013). Conversion of gamma-glutamylanilide into catechol is the next step which is catalyzed by aniline dioxygenases. High concentrations of gamma-glutamylanilide are considered cytotoxic agent, but the action of glutamine amidotransferase enzyme prevents its accumulation by converting it to aniline (Takeo et al., 2013). These three enzymes are encoded by three genes which have been identified in some bacteria like P. putida UCC22 (Fukumori and Saint, 1997), Acinetobacter spp.YAA (Fujii et al., 1997), Delftia acidovorans 7N (Urata et al., 2004), Delftia tsuruhatensis AD9 (Liang et al., 2005), Frateuria sp.ANA-18(Murakami et al., 2003) and Delftia sp.AN3 (Zhang et al., 2008). These genes are carried on either chromosomal DNA or plasmid. The plasmids of Acinetobacter spp.YAA (pYA1) and P. putida UCC22 (pTDN1) contain aniline oxidation genes (atdA1A2A3A4 or tdnQTA1A2B)(Fujii et al., 1997; Fukumori and Saint, 1997). Also all five genes from Frateuria sp.ANA-18 were expressed in Escherichia coli, and their recombinant bacteria exhibited the aniline oxidation activities (Murakami et al., 2003). It was demonstrated that loss of activity of aniline oxidation is was the result of deletion of tdnA1A2 or tdnQ genes.
The aim of the present study was isolation, identification and molecular characterization of bacterial strains capable of degrading aniline, in addition to studying the genetic basis of biodegradation.
Aniline of analytical grade was purchased from El Nasr (Egypt). HPLC-grade acetonitrile and water were purchased from LAB-Scan (Poland). Presterilized polystyrene; flat-bottom, tissue culture- 96-well microtiter plates (Corning Incorporated, Corning, N.Y.). Oxidase reagent was purchased from Difco, USA. All other chemicals were of analytical grade.
All media used for isolate identification (Triple sugar iron agar, MacConKey agar medium, cetrimide agar medium, lysine iron agar medium, Motility ornithine indole medium, Tryptone Soya broth, Tryptone Soya agar, and Simmons citrate agar medium) were purchased from Oxoid, UK.
A mineral salt medium (MSM) (pH 7.0 ± 0.1) contained (g/l deionized water), K2HPO4 2; KH2PO4 1; NaNO3 4; CaCl2 0.1; KCl 1; NaCl 1; MgSO4.7H2O 0.2 and 1 ml of Focht trace elements solution (Focht, 1994). Focht trace elements solution contains (mg/l) MnSO4.H2O 169; ZnSO4.7H2O 288; CuSO4.5H2O 250; NiSO4.6H2O 26; CoSO4 28 and NaMoO4.2H2O 24 (pH 7.2 ± 0.2). The above liquid media were solidified by adding 2% noble agar (w/v) (Difco, USA). Luria-Bertani (LB) medium composition was (g/l deionized water): Tryptone 10.0; yeast extract 5.0 and NaCl 10.0 (pH 7.0 ± 0.2).
Isolation and biochemical identification of aniline degrading bacteria
Thirty agriculture wastewater and domestic sewage samples were collected from different sites from Beni-Suef City, Egypt. Nearly 50-100 ml of each sample was collected in 250 ml sterile conical flasks that were stored at 4°C till use. Aniline was dissolved in acetone and sterilized by membrane filtration using a 0.22 µm Millipore filter. MSM was autoclaved, cooled to 40°C and then supplemented with the required concentration of aniline. Ten milliliters of each sample was added to 90 ml of MSM supplied with aniline at a concentration of 100 mg/l as a sole source of carbon and energy, incubated at 30°C in a rotary shaker (REMI, India) at150 rpm for 10 days. After 3 successive transfers, the culture was serially diluted and spread on MSM plates containing 100 mg/l of aniline, incubated for 7 days at 30°C. Discrete, isolated colonies based on morphological properties were selected and purified to obtain a pure culture.
These isolates were identified depending on Gram stain, cultures characters of colonies, growth on MacConkey agar, growth on cetrimide agar, catalase test, oxidase test, gelatinase test, citrate utilization test, motility test, ornithine decarboxylation test, the reaction on lysine iron agar, indole test and reaction on triple sugar iron agar. All media were purchased from Oxoid, UK.
DNA isolation and manipulation
The genomic DNA of the isolated microorganisms was extracted using GenoElute DNA Bacterial Genomic DNA Kit (Cat. no NA2110) (Sigma-Aldrich, USA). The plasmid of each isolate was isolated using the Qiaprep Spin Miniprep kit (Cat. no 27104) (Qiagen, Germany). The kits were used according to the instructions of the manufacturers.
The success of genomic DNA and plasmid extraction was confirmed by running DNA extracts on 0.8% agarose gel (w/v) (Invitrogen) stained with ethidium bromide (Thermo Fischer Scientific, USA) using gel electrophoresis (Labnet, USA). Then, the gel is illuminated under Whatman U.V Transilluminator (Biometra, Germany) according to (Sambrook et al., 1989). DNA concentration and purity were determined using the Nano-drop 2000- spectrophotometer (Thermo Fischer Scientific, USA), as ng per µl for concentration and by dividing 260/280 nm UV absorbance readings to obtain purity.
Identification of Aniline-degrading isolates by 16S rRNA
Identification was confirmed by sequencing of partially amplified 16S rRNA gene of the bacterial isolate. Molecular characterization of the selected strains was carried out according to modified (Sambrook et al., 1989).
For isolate 1 and isolate 4, the 16S rRNA gene was amplified using universal primers pair: V1 Fw (5-AGAGTTTGATCATGGCTCAG-3) and V6 Rv (5-ACGAGCTGACGACAGCCATG-3) (IDT, USA) recorded to produce an amplicon of approximately 1073 base pair (bp)(Sundquist et al., 2007). For isolates 2, 3 and 5, 16S rRNA gene was amplified using FD1 (5-CCGAATTCG TCGACAAC AGAGTTTG ATCCTGGCTCAG -3), and RP1 (5-CCCGGGATCCAAGCTT ACGGTTACCTT GTTACGACTT -3) (IDT, USA) recorded to produce an amplicon of approximately 1400 base pair (bp).
Primers solutions were prepared according to the instructions of the manufacturer to a concentration of 100 µM stock solution using nuclease-free water.
PCR was carried out in a total volume of 50 ml using 25 ml of MyTaq PCR Master Mix (2X) (Bioline, Germany), containing 1 ml (10 mM) of each forward and reverse primers together with equivalent microliters to 50 ng of the genomic DNA and finally completed to 50 ml by nuclease-free water. Thermocycling was carried out using thermocycler (Primus 25 Advanced, Germany), the PCR amplification of the 16S rRNA sequences was done according to the thermal profile shown in the table (1).
Table (1):
The used thermal profile for the amplification of 16S rRNA gene by PCR.
| Conditions | Temperature | Time | ||
|---|---|---|---|---|
| Steps | ||||
| 1 | Initial denaturation | 94oC | 3 min | |
| 2 |
40 Cycles |
Denaturation | 94oC | 30 sec |
| 3 | Annealing | 50oC for V1 Fw & V6 Rv 56oC for FD1 & RP1 | 30 sec | |
| 4 | Extension | 72oC | 2 min | |
| 5 | Final extension | 72oC | 10 min. | |
The amplification was verified by gel electrophoresis using 1% (w/v) agarose gel supplemented with 0.2 mg/ml of ethidium bromide. PCR products were purified by Gene Jet Gel Extraction Kit (Thermo Fisher Scientific, USA) according to manufacturer protocol. The purified products were sent for sequencing at Macrogen, Korea. Nucleotide sequence similarities were determined using other known sequences found in the GenBank database using the BLAST program of National Center for Biotechnology Information (NCBI) databases.
Inoculum preparation for Aniline biodegradation study
Inoculum size of highly aniline degrader was prepared as described by(Anwar et al., 2009). Briefly, a single pure colony from the strain was grown in nutrient broth medium, harvested by centrifugation at 4600×g for 5 min, washed with autoclaved normal saline and resuspended in normal saline to set an OD620 nm of 0.08. Colony forming units per milliliter (CFU/ml) of this suspension was quantified by the dilution plate count technique and required inoculums be prepared by adding an appropriate amount of normal saline.
Analytical Method
One hundred milliliters of MSM supplemented with 100 mg/l of aniline was inoculated with bacterial cells to obtain a concentration of 1.5×108 CFU/ml and incubated at 30°C in an orbital shaker at 150 rpm. Aniline residues were detected at different time intervals 0, 2, 4, 6, 8 and 10 days from inoculation. Each treatment was performed in three replicates. A set of media without microorganism amended with an equal amount of aniline was run simultaneously under identical conditions.
Samples used for HPLC analysis were prepared by centrifugation at 10,000g for 15 min, and the resulting supernatants were syringe filtered through 0.22 µm nylon filters. Five milliliters aliquot from the previously mentioned different media were centrifuged at 7200×g for 10 min to remove the bacterial pellets and extracted two times with 50 ml of dichloromethane by shaking vigorously. Dichloromethane extracts were then passed through anhydrous Na2SO3 for dehydration and evaporated to dryness using rotary evaporator at 30°C. The dry residue was dissolved in 1 ml acetonitrile and subjected to HPLC analysis. Twenty microliters were injected in HPLC (Agilent Technologies 1260 Infinity, USA), using Eclipse plus C18 (4.6 ×250mm 5 µm) and diode array detector (DAD) at l 230 nm. HPLC working conditions were acetonitrile: water (70:30), at a flow rate of 1 ml/min. Aniline concentration was calculated by comparison to a standard curve.
The metabolic pathway of aniline biodegradation was also investigated using the HPLC technique. The formation of catechol as a byproduct of aniline biodegradation was detected at 270nm. Also, further cleavage of catechol by catechol 1,2 dioxygenase or catechol 2,3 dioxygenase was detected by formation of cis,cis-muconic acid at 260nm or muconic acid semialdehyde at 375 nm respectively.
Detection of isolate growth by optical density method
The bacterial cell mass was determined by transferring aseptically 200 microliters of samples to a microtiter plate and measuring the optical density at 620 nm (OD620) using microtiter plate reader (Sunrise, Tecan, Austria). The experiment was performed in triplicate.
Plasmid curing
The isolates (about 105 cfu ml-1) were inoculated in tryptone soya broth with different concentrations of the curing agent (1-32 µg ml-1 ethidium bromide, Fischer Scientific) and incubated for 18 h at 30°C. Cultures that displayed growth at the highest concentration of ethidium bromide were serially diluted and plated onto tryptone soya agar to obtain isolated colonies (15-20 per plate) (Sabia et al.). After 24 h of incubation at 30°C, each colony was picked up and inoculated on Noble agar plates containing aniline as the sole carbon source. The loss of aniline biodegradation activity was indicated by a failure of the transferred colonies to grow on Noble agar plates which are an indication of plasmid curing. The success of plasmid curing was also confirmed by plasmid extraction.
PCR amplification of specific sequences of some genes encoding for aniline biodegradation
Four genes contributing to aniline biodegradation in all isolates were selected in this study. These selected genes were cat 1,2, cat 2,3, tdnQ and tadR genes. These genes were amplified by PCR using primers in the table (2). All genes were screened in both genomic DNA and plasmid preparation.
Table (2):
List of primer pairs used in the study, their sequences, their expected product size and their references.
Reference |
Product Size |
Sequence |
Primer |
Gene |
|---|---|---|---|---|
This Study |
203 |
F (5-CAAGGTGCTCGGCTTCTATC-3) R (5-GTCGGTCATGGAGATCAGGT-3) |
Forward Reverse |
cat 2,3 |
(Civilini et al., 1999) |
605 |
F (5-AGACCTGGAAATCACCGAAG ACG-3) R (5-GGGTGGCGTAGGCAAAGTC GTC-3) |
Forward Reverse |
cat 1,2 |
(Shin and Spain, 2009) |
370 |
F (5-GGCTGGATGCTGGCAGATCT-3) R (5-AAGGTGGTTTCCATCTGGCT-3) |
Forward Reverse |
tdnQ |
(Geng et al., 2009) |
1407 |
F (5-GATGGATCCGAGGCGCTGCTC-3) R (5-GCTCTAGACGATGCGCACCTTG-3) |
Forward Reverse |
tadR |
The sequence of cat 2,3 gene was taken from the GenBank sequence database of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/). These sequences were aligned by the CLC sequence analyzer program and the Multiple Sequence Alignment (SRS) using the online program(http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-page+Launch+-id+12F2q1huXNL+-appl+NClustalW2+-launchFrom+top) to detect the conserved region of that gene. PCR primers of this gene were designed with the online program primer 3 (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) available by IDT, USA.
The PCR reaction was performed in a total volume of 25 ml containing 12.5 µl of MyTaq PCR Master Mix (2X) (Bioline, Germany), 1 ml of each forward and reverse primer (10 mM) together with equivalent µls to 50 ng of the genomic DNA and finally completed to 25 ml by nuclease-free water. Amplification was performed using Primus 25 Advanced, Germany thermocycler according to thermal profile in the table (3).
Table (3):
The thermal profile used for the screening of aniline biodegradation genes by PCR.
| Condition | Temperature | Time | ||
|---|---|---|---|---|
| Steps | ||||
| 1 | Initial denaturation | 94°C | 3 min | |
| 2 | Denaturation | 35 cycles | 94°C | 30 sec |
| 3 | Annealing | 54°C for tdnQ 52°C for cat 2,3 57.5 for tadR |
30 sec | |
| 4 | Extension | 72°C | 2 min | |
| 5 | Final extension | 72°C | 10 min | |
The PCR reaction for cat 1,2 was performed according to protocol; 94°C for 3 min followed by 25 cycles of 94°C for 30 sec, 65°C for 1 min and 72°C for 1 min, followed by 15 cycles at 94°C for 30 sec, 55°C for 1 min and 72°C for 1 min with a final extension at 72°C for 10 min.
Finally, the PCR products were detected by gel electrophoresis by running on 1.5% agarose gel (w/v) stained with ethidium bromide using 100 bp DNA ladder (Bioline, Germany) as a reference and then illuminated under U.V spectrophotometer according to(Sambrook et al., 1989).
Isolation and identification of aniline degrading bacteria
Collected samples were subcultured on the liquid medium of MSM supplied with 100 mg/l of aniline.
The present study was conducted on five isolates which their growth, biochemical and morphological characteristics supported its identity being P. aeruginosa 1 (Isolate No. 1), A. cholinophagum (Isolate No. 2), E. asburiae (Isolate No. 3), P. aeruginosa 2 (Isolate No. 4), A. xylosoxidans (Isolate No. 5). Bacterial identification using 16S rRNA sequence analysis confirmed the results of the biochemical tests. The sequences were compared to GenBank sequence database of NCBI for analysis using the online program http://blast.ncbi.nlm.nih.gov/Blast.cgi. Sequence blast showed that the isolate No. (1) had a high similarity 99 % to P. aeruginosa strain KS3 (KF021246.1), 99 % similarity to P. aeruginosa strain Z11 (AY548953.1). While isolating No. (2) had a high similarity 99 % to A. cholinophagum strain NBRC 102400 (AB681756.1), 85% similarity to A. xylosoxidans strain RE1 (FM163487.1 ). The isolate No. (3) had a high similarity 99 % to E. asburiae strain IHB B 1070 (KF475833.1), 92 % similarity to E. aerogenes strain Z93 (KF835769.1). For isolate No. (4), it had a high similarity of 100 % to P. aeruginosa strain LWIS(KT945245.1), 100 % similarity to P. aeruginosa strain YJY3 (KP973963.1. For isolate No. (5), it had a high similarity 100 % to A. xylosoxidans isolate wp22 (AJ560626.1), 86 % similarity to A. xylosoxidans strain yb97 (EU214610.1).
Detection of isolate growth by optical density method
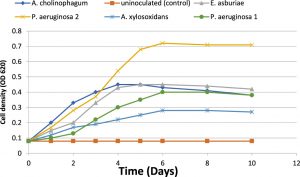
The growth kinetics of the five isolates were investigated by growing in a minimal medium of 100 mg/L of aniline concentration. The growth curve of the five isolates in the presence of aniline was determined by the measurement of the optical density over 10 days of incubation. The growth curves showed that all isolates had rapid growth in the presence of aniline. The isolates reached the maximum optical density from the second to the sixth day of incubation. The growth curves showed an ascendant situation in the first six days, and finally, the bacterial growth rate was decreased (Figure 1).
Fig. 1. Microbial growth (cell density) of the five isolates during aniline biodegradation using it as a sole carbon source (f&) and uninoculated (control) (%).
HPLC analysis of the aniline residue
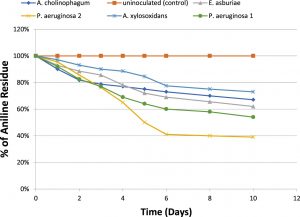
For determination of the degradative capacity of the five isolates, MSM containing 100 mg/l of aniline as a sole source of carbon and energy were inoculated with 1.5×108 cfu ml-1 of the previously mentioned bacterial isolates separately. They were then incubated at 150 rpm for 10 days. Aniline degradation was monitored throughout the 10 days using HPLC. At the end of incubation, only 46% of aniline was degraded by P.aeruginosa (1), 33% of aniline was degraded by A. cholinophagum, and 38% of aniline was degraded by E. asburiae. On the other hand, P. aeruginosa (2) was the most efficient degrading bacterium with 61% degradation of aniline, while, A. xylosoxidans was the least efficient degrading bacterium with 27% degradation of aniline (Figure 2).
Fig. 2. Percentage of aniline biodegradation by the five isolates in MSM containing aniline (f&) at a concentration of 100 mg /l in MSM using aniline as the sole carbon and energy source, and uninoculated (control) (%).
Identification of metabolites produced from aniline biodegradation
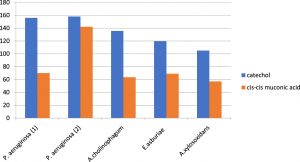
Metabolic pathway of aniline biodegradation was also investigated in this study by using the HPLC chromatography. It was observed that catechol was the first byproduct produced by aniline degradation and then it was further biodegraded to cis, cis- muconic acid following the ortho cleavage pathway. Catechol was first produced by all isolates reaching its peak early. However, the onset of formation of cis, cis- muconic acid was slowly reaching its peak later on. Both of them were increasing exponentially as aniline concentration decreased and finally leveled off (Figure 3).
Fig. 3. Formation of different metabolites as byproducts of aniline degradation by the five aniline degrading isolates.
Plasmid extraction
The success of plasmid extraction of the five selected isolates was confirmed by running the DNA extracts on 1% agarose gel electrophoresis. The concentration and the purity of the isolated plasmids were estimated using the Nanodrop spectrophotometer. The Nanodrop spectrophotometer results indicated that DNA concentrations of the extracts were variable ranging from 38.3 ng /µL to 160.7 ng/ µL It was also observed that purity of DNA extracts was high ranging from 1.83 to 1.88
PCR screening of the selected genes
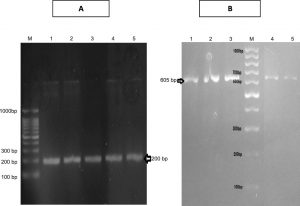
PCR screening followed by gel electrophoresis for the specific sequences of the four selected genes, cat 2,3, cat 1,2, tdnQ and tadR, were done for the studied five isolates. It revealed the presence of cat 1,2 and cat 2,3 genes in both the extracted genomic as well as plasmid DNA, resulted in amplification of 605 bp product of cat1,2 gene and 203 base pair of cat2,3 gene (Figures 4).
Fig. 4. A) Agarose gel electrophoresis of amplicon of specific sequence of cat2,3 gene (200 bp) in all isolates . B) Agarose gel electrophoresis of amplicon of specific sequence of cat1,2 gene (605 bp) in all isolates.
Regarding tdnQ gene detection, a PCR product was observed at a size of 370 bp in its PCR amplification. This gene was detected in both the extracted genomic as well as plasmid DNA of isolates No.1 & 4.
Regarding tadR gene detection, a PCR product was observed at 200 bp upon PCR. This gene (regulatory gene) was detected in the screened four isolates (isolates No.1, 2, 4 & 5) in both the extracted genomic as well as plasmid DNA.
Plasmid curing
Plasmid curing was performed in order to determine the involvement of plasmid-encoded genes in aniline degradation, . It was observed that some colonies were unable to utilize aniline and thus failed to grow and this was due to the success of the plasmid curing. The success of curing was confirmed by the extraction of plasmids and the products obtained were electrophoresed. The result indicated that no plasmids were extracted from plasmid-cured isolates. So, it was concluded that the genes involved in aniline degradation were plasmid encoded.
In this study, thirty domestic sewage and agriculture wastewater samples and soil samples were subcultured on the liquid medium of MSM supplied with 100 mg L-1 of aniline as a sole carbon source. A total of five bacterial isolates were recovered from the different samples using the low concentration of aniline. Plating of samples on MSM agar of aniline, isolates were identified as P. aeruginosa (1 and 2), A. cholinophagum, A. xylosoxidans, and E. asburiae according to the biochemical identification and 16s RNA result analysis.
Previous studies reported that several bacterial genera such as; Delftia sp. AN3 (Liu et al., 2002), Rhodococcus sp. AN5 (Zhuang et al., 2007), Acinetobacter spp.YAA (Fujii et al., 1997), Erwinia sp. HSA 6 (Li et al., 2010) and Comamonas testosteroni I2 (Boon et al., 2000) had the ability of aniline biodegradation. Also, Pseudomonas putida UCC22 (Fukumori and Saint, 1997), Pseudomonas sp.AW- 2 (Murakami et al., 1998), and Delftiat suruhatensis AD9 (Liang et al., 2005) may participate in efficient aniline biodegradation. In 1992, Six bacterial strains, able to degrade aniline, were isolated belonging to Pseudomonas putida, Pseudomonas acidovorans, Achromobacter gr. D.V, Achromobacter xylosoxidans, Moraxella sp. K21 and Moraxella sp. K22 (Kahng et al., 1992).
(Kahng et al., 2000b) isolated aniline degrading microorganisms from the investigated soil in Sothern Korea. Aniline degrading bacteria were identified via usual biochemical tests and PCR method with the 16S rRNA gene (Kafilzadeh et al., 2013). In our study, the growth kinetics of the five isolates was investigated with 100 mg/L of aniline. Growth curve of isolates in the presence of aniline during 10 days of incubation is shown in Figure (1). The five isolates showed rapid growth in the presence of aniline. The growth curve showed an ascendant situation, where the isolate reached the maximum optical density from the second to the sixth day of incubation. Eventually, the bacterial growth rate was decreased with time as a result of aniline depletion as a sole carbon source and also increase the toxicity of secondary metabolites.
In a similar study of aniline biodegradation (Kafilzadeh et al., 2013), they investigated the growth kinetics of Pseudoalteromonas arctica and Corynebacterium sp. via OD method, where bacteria reached to the maximum optical density from the third to the sixth day of incubation then decreasing in growth rate with time was observed. On the other hand, the growth kinetics of Pseudomonas acidovorans and achromobacter sp. showed that the optical density reached to a maximum from 24 to 36 hours then decreased after 50 to 72 hours (Kahng et al., 1992).
The degradative capacity of the five isolates was determined by using HPLC. The five isolates were able to grow on high aniline concentration reflecting their ability to use it as a sole source of carbon and energy. We found that about 46% of aniline was degraded by P. aeruginosa 1, about 61% by P. aeruginosa 2, 33% by A. cholinophagum, 38% by E. asburiae and 27% by A. xylosoxidans. Within 10 days of incubation indicated by analysis of the remaining concentrations of aniline using HPLC. Similarly, in another study, about 50% of aniline was degraded by Aspergillus ustus within 7 days(Hatvani et al., 2013). Also, it was previously reported that about 31.23% of aniline was degraded by Staphylococcus aureus ST1, after incubation for three days(Ahmed et al., 2009), however, (Liu et al., 2002) showed that Delftia sp. AN3 degraded aniline by 100% within 7 days.
The available literature data showed that biodegradation of aniline in various bacterial cultures might proceed either according to extradiolic (meta-cleavage) or intradiolic (ortho-cleavage) pathway. In both cases, the primary intermediate of aniline biodegradation is catechol (Mucha et al., 2009).
Colonies of the five isolates cultured on plates or suspended in MSM media did not develop a yellow colour.W indicates that there was no formation of 2 hydroxymuconic semialdehyde of the meta-cleavage pathway(Kim and Zylstra, 1995)i. It was then assumed that the isolates probably might metabolize aniline via the ortho-cleavage pathway. Then the next step was to perform the HPLC chromatography method to detect the presence of catechol and cis, cis muconic acid, the intermediate indicator for cleavage pathway. HPLC results revealed that the five isolates biodegraded aniline according to the ortho-cleavage pathway resulting in cleavage of the benzene ring of the catechol byproduct into cis, cis muconic acid.
It was observed that catechol was first produced in all isolates reaching its peak early; however, the onset of formation of cis, cis muconic acid was slowly reaching its peak later on. Both of them were increasing exponentially as aniline concentration decreased and finally leveled off. The concentration of catechol was found to be growing up to the 5th day then the onset of formation of cis, cis muconic was after 48 hours of the degradation process in all isolates except for P.aerugenosa 2 where it was formed after 24 hours due to high the degradation power of the organism and increased exponentially as aniline concentration decreased. Finally, it leveled off after 10 days incubation period and appeared to accumulate in the medium.
It was previously reported that the degradation of chloroanilines occurs via a dioxygenation and deamination, resulting in the most likely intermediate product, chlorocatechol (Radianingtyas et al., 2003; Zeyer et al., 1985). Also, the result of HPLC used in the biodegradation of aniline by Candida methanosorbosa BP-6 showed that catechol concentration was found to be growing up to the day 5, and the level of cis, cis muconic acid was highest between days 3 and 5 of fermentation which was similar to our results (Mucha et al., 2010).
On the other hand, the onset of formation of cis, cis muconic acid was after 6 hours of the degradation process of phenol by Candida. tropicalis RETL-Cr1 (AY725426) which seemed to metabolize phenol via ortho-cleavage pathway (Tuah et al., 2004).
Also, the application of specific primers for cat1,2 and cat 2,3 genes encoding for production of catechol 1,2 dioxygenase and catechol 2,3 dioxygenase enzymes respectively, resulted in amplification of 203 base pair of cat2,3 gene and 605 bp product of cat1,2. These results confirmed that aniline was degraded by the ortho-cleavage pathway of the resulting catechol due to the detection of for cat1,2 gene encoding for production of catechol 1,2 dioxygenase which is responsible for the ortho-cleavage pathway. However, the presence of cat2, 3 gene, and absence of its byproduct (2 hydroxymuconic semialdehyde) made us suggesting that this gene not is expressed in the isolates due to the absence of 2 hydroxymuconic semialdehyde on HPLC result.
Frateuria sp. ANA18 (Murakami et al., 2003), Delftia sp. XYJ6 (Chengbin et al., 2009) and Candida methanosorbosa BP-6 showed to degrade aniline via the ortho-cleavage pathway with the formation of cis, cis muconic acid. However, Acinetobacter spp.YAA (Fujii et al., 1997) and Delftia tsuruhatensis AD9 (Liang et al., 2005) were reported to metabolize aniline via the meta-cleavage pathway with the formation of 2-hydroxymuconic semialdehyde.
In the current study, also tdnQ gene was detected by PCR yielding a product of 370 base pair from the P. aeruginosa (1 and 2) only. Similarly, the tdnQ gene was found in P. putida (Fukumori and Saint, 1997). While tadQ gene and atdA gene (aniline oxygenase encoding gene which has the same function of tdnQ) were found in Delftia tsuruhatensis AD9 and Acinetobacter spp. YAA strain, respectively (Geng et al., 2009).
Regarding tadR gene detection, a PCR product was observed at 1407 bp in its PCR amplification. This gene (regulatory gene) was detected in only four isolates (P. aeruginosa (1 and 2), A. cholinophagum and A. xylosoxidans in both the extracted genomic DNA as well as plasmids, while it was not detected in E. asburiae isolate. The gene product of tadR (TadR) showed particularly high identity with putative LTTRs for aniline degradation [TdnR from P. putida UCC22 97.6% (BAA12810) (Geng et al., 2009).
The bacterial transformation of aromatic compounds has frequently been shown to be plasmid encoded as reported for many microbial strains (Anson and Mackinnon, 1984; Saint et al., 1990).
Before isolation of plasmid having susceptible degradation genes, plasmid curing suggested by (Mäe et al., 1993) was performed to ensure that the gene degrading aniline is plasmid encoded or chromosomal encoded. Plasmid curing of the five isolates showed that 90% of P. aeruginosa (1), 75% of A. cholinophagum, 60% of E. asburiae, 85% of P. aeruginosa (2) and 64% of A. xylosoxidans colonies failed to grow on aniline agar plates which were possibly due to fact that genes involved in aniline catabolic pathway were plasmid encoded. Also, this result was confirmed by the absence of plasmids bands on the gel after extraction of plasmids from the cured bacteria.
Similarly to our results, 90% of Staphylococcus aureus ST1 colonies founded to be unable to grow on aniline agar plates [55]. Also, 36% of Acinetobacter sp. strain YAA colonies could not grow on aniline on using acridine orange mutagenicity (Fujii et al., 1997).
The plasmids of P. putida UCC22 (pTDN1) and Acinetobacter spp.YAA (pYA1) containing aniline oxidation genes, tdnQTA1A2B or atdA1A2A3A4 (Fujii et al., 1997; Fukumori and Saint, 1997) were fully sequenced in another study. However, in another study by (Liang et al., 2005) aniline biodegradation cluster (tadQTA1A2B) was detected in chromosomal DNA of Delftia tsuruhatensis AD9.
Our isolated bacteria could degrade aniline as a sole carbon source, and we founded that our strains were belonging to Pseudomonas, Enterobacter and achromobacter genera could degrade aniline via an ortho cleavage pathway. So, these isolates should be used in aniline biodegradation.
However, the presence of cat 2,3 gene and absence of its byproduct (2 hydroxymuconic semialdehyde) made us suggesting that this gene is expressed in the isolates due to the absence of 2 hydroxymuconic semialdehyde on HPLC result. Additionally, genes responsible for aniline biodegradation were detected in the isolates, and they have located on plasmid only not the chromosome. E. asburiae was not very common microorganism in aniline biodegradation as it was not studied intensely in other research articles of aniline biodegradation.
- Ahmed, S., Nisar, M., Hussain, K., Majeed, A., Ghumroo, P., Afghan, S., Shahzad, A., Nawaz, K. and Ali, K. Isolation and characterization of a bacterial strain for aniline degradation. African Journal of Biotechnology, 2009; 9:1173-1179
- Anson, J. G. and Mackinnon, G. Novel Pseudomonas plasmid involved in aniline degradation. Applied and Environmental Microbiology, 1984; 48: 868-869.
- Anwar, S., Liaquat, F., Khan, Q. M., Khalid, Z. M. and Iqbal, S. Biodegradation of chlorpyrifos and its hydrolysis product 3, 5, 6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. Journal of Hazardous Materials, 2009; 168: 400-405.
- Aoki, K., Ohtsuka, K., Shinke, R. and Nishira, H. Rapid biodegradation of aniline by Frateuria species ANA-18 and its aniline metabolism. Agricultural and biological chemistry, 1984; 48: 865-872.
- Bomhard, E. M. and Herbold, B. A. Genotoxic activities of aniline and its metabolites and their relationship to the carcinogenicity of aniline in the spleen of rats. Critical reviews in toxicology, 2005; 35: 783-835.
- Boon, N., Goris, J., De Vos, P., Verstraete, W. and Top, E. M. Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, I2gfp. Applied and Environmental Microbiology , 2000; 66: 2906-2913.
- Boon, N., Goris, J., De Vos, P., Verstraete, W. and Top, E. M. Genetic Diversity among 3-Chloroaniline-and Aniline-Degrading Strains of theComamonadaceae. Applied and Environmental Microbiology, 2001; 67:1107-1115.
- Bull, R. J., Reckhow, D. A., Li, X., Humpage, A. R., Joll, C. and Hrudey, S. E. Potential carcinogenic hazards of non-regulated disinfection by-products: haloquinones, halo-cyclopentene and cyclohexene derivatives, N-halamines, halonitriles, and heterocyclic amines. Toxicology, 2011; 286: 1-19.
- Carmona, M., Zamarro, M. T., Blzquez, B., Durante-Rodrguez, G., Jurez, J. F., Valderrama, J. A., Barragn, M. J., Garca, J. L. and Daz, E. Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiology and Molecular Biology Reviews, 2009; 73: 71-133.
- Chengbin, X., Jun, N., Hai, Y., Xudong, S. and Jiye, H. Biodegradation of aniline by a newly isolated Delftia sp. XYJ6. Chinese Journal of Chemical Engineering, 2009; 17: 500-505.
- Civilini, M., Bertoldi, E. and Tell, G. Molecular characterization of Pseudomonas aeruginosa 2NR degrading naphthalene. Letters in applied microbiology, 1999; 29: 181-186.
- DeBruin, L. S. and Josephy, P. D. Perspectives on the chemical etiology of breast cancer. Environmental health perspectives, 2002; 1: 119, 128.
- Essington, M. E. Adsorption of aniline and toluidines on montmorillonite. Soil science, 1994; 158: 181-188.
- Fujii, T., Takeo, M. and Maeda, Y. Plasmid-encoded genes specifying aniline oxidation from Acinetobacter sp. strain YAA. Microbiology, 1997; 143: 93-99.
- Fukumori, F. and Saint, C. P. Nucleotide sequences and regulational analysis of genes involved in conversion of aniline to catechol in Pseudomonas putida UCC22 (pTDN1). Journal of bacteriology, 1997; 179: 399-408.
- Geng, L., Chen, M., Liang, Q., Liu, W., Zhang, W., Ping, S., Lu, W., Yan, Y., Wang, W. and Takeo, M. Functional analysis of a putative regulatory gene, tadR, involved in aniline degradation in Delftia tsuruhatensis AD9. Archives of microbiology, 2009; 191: 603-614.
- Hatvani, L., Manczinger, L., Marik, T., Bajkán, S., Vidács, L., Bencsik, O., Szekeres, A., Radulov, I., Nita, L. and Vágvölgyi, C. The complete degradation of acetanilide by a consortium of microbes isolated from River Maros. Acta Biologica Szegediensis, 2013; 57: 117-120.
- Kafilzadeh, F., Nejad, R. A. and Nejad, F. M. Isolation and identification of aniline degrading bacteria from sediments of Kharg island in Persian Gulf. European Journal of Experimental Biology, 2013; 3:454-460.
- Kahng, H. Y., Kim, S. I., Woo, M. J., Park, Y. K. and Lee, Y. N. Isolation and characterization of aniline-degrading bacteria. The Korean Journal of Microbiology, 1992; 30: 199-206.
- Kahng, H. Y., Kukor, J. J. and Oh, K. H. Characterization of strain HY99, a novel microorganism capable of aerobic and anaerobic degradation of aniline. FEMS microbiology letters, 2000a; 190: 215-221.
- Kahng, H. Y., Kukor, J. J. and Oh, K. H. Physiological and phylogenetic analysis of Burkholderia sp. HY1 capable of aniline degradation. Journal of microbiology and biotechnology, 2000b; 10: 643-650.
- Kim, E. and Zylstra, G. J. Molecular and biochemical characterization of two meta-cleavage dioxygenases involved in biphenyl and m-xylene degradation by Beijerinckia sp. strain B1. Journal of bacteriology, 1995; 177: 3095-3103.
- Li, J., Jin, Z. and Yu, B. Isolation and characterization of aniline degradation slightly halophilic bacterium, Erwinia sp. Strain HSA 6. Microbiological research, 2010; 165:418-426.
- Liang, Q., Takeo, M., Chen, M., Zhang, W., Xu, Y. and Lin, M. Chromosome-encoded gene cluster for the metabolic pathway that converts aniline to TCA-cycle intermediates in Delftia tsuruhatensis AD9. Microbiology, 2005; 151: 3435-3446.
- Liu, Z., Yang, H., Huang, Z., Zhou, P. and Liu, S. J. Degradation of aniline by newly isolated, extremely aniline-tolerant Delftia sp. AN3. Applied microbiology and biotechnology, 2002; 58: 679-682.
- Loidl, M., Hinteregger, C., Ditzelmller, G., Ferschl, A. and Streichsbier, F. Degradation of aniline and monochlorinated anilines by soil-born Pseudomonas acidovorans strains. Archives of microbiology, 1990; 155: 56-61.
- Mäe, A. A., Marits, R. O., Ausmees, N. R., Kôiv, V. M. and Heinaru, A. L. (). Characterization of a new 2, 4-dichlorophenoxyacetic acid degrading plasmid pEST4011: physical map and localization of catabolic genes. Microbiology, 1993; 139: 3165-3170.
- Markowitz, S. B. and Levin, K. Continued epidemic of bladder cancer in workers exposed to ortho-toluidine in a chemical factory. Journal of occupational and environmental medicine, 2004; 46:154-160.
- Mucha, K., Kwapisz, E., Kucharska, U. and Okruszek, A. Mechanism of aniline degradation by yeast strain Candida methanosorbosa BP-6. Pol J Microbiol, 2009; 59: 311-315.
- Murakami, S., Hayashi, T., Maeda, T., Takenaka, S. and Aoki, K. Cloning and functional analysis of aniline dioxygenase gene cluster, from Frateuria species ANA-18, that metabolizes aniline via an ortho-cleavage pathway of catechol. Bioscience, biotechnology, and biochemistry, 2003; 67: 2351-2358.
- Murakami, S., Nakanishi, Y., Kodama, N., Takenaka, S., Shinke, R. and Aoki, K. Purification, characterization, and gene analysis of catechol 2, 3-dioxygenase from the aniline-assimilating bacterium Pseudomonas species AW-2. Bioscience, biotechnology, and biochemistry, 1998; 62: 747-752.
- Palmiotto, G., Pieraccini, G., Moneti, G. and Dolara, P. Determination of the levels of aromatic amines in indoor and outdoor air in Italy. Chemosphere, 2001; 43: 355-361.
- Pluvinage, B., Dairou, J., Possot, O. M., Martins, M., Fouet, A. S., Dupret, J. M. and Rodrigues-Lima, F. Cloning and molecular characterization of three arylamine N-acetyltransferase genes from Bacillus anthracis: identification of unusual enzymatic properties and their contribution to sulfamethoxazole resistance. Biochemistry, 2007; 46: 7069-7078.
- Radianingtyas, H., Robinson, G. K. and Bull, A. T. Characterization of a soil-derived bacterial consortium degrading 4-chloroaniline. Microbiology, 2003; 149: 3279-3287 .
- Rai, H. S., Bhattacharyya, M. S., Singh, J., Bansal, T., Vats, P. and Banerjee, U. Removal of dyes from the effluent of textile and dyestuff manufacturing industry: a review of emerging techniques with reference to biological treatment. Critical reviews in environmental science and technology, 2005; 35: 219-238.
- Sabia, C., Anacarso, I., Bergonzini, A., Gargiulo, R., Sarti, M., Condò, C., Messi, P., De niederhausern, S., Iseppi, R. and Bondi, M. Detection and partial characterization of a bacteriocin-like substance produced by Lactobacillus fermentum CS57 isolated from human vaginal secretions. Anaerobe, 2014; 26: 41-45.
- Saint, C. P., McClure, N. C. and Venables, W. Physical map of the aromatic amine and m-toluate catabolic plasmid pTDN1 in Pseudomonas putida: location of a unique meta-cleavage pathway. Microbiology, 1990; 136: 615-625.
- Sambrook, J., Fritsch, E. and Maniatis, T. 1989. Cold Spring Harbour Laboratory Press. NY, USA 23.
- Shin, K. A. and Spain, J. C. Pathway and evolutionary implications of diphenylamine biodegradation by Burkholderia sp. strain JS667. Applied and environmental microbiology, 2009; 75: 2694-2704.
- Sihtmae, M., Mortimer, M., Kahru, A. and Blinova, I.Toxicity of five anilines to crustaceans, protozoa and bacteria. Journal of the Serbian Chemical Society, 2010; 75: 1291-1302.
- Stellman, J. M. Encyclopaedia of occupational health and safety, 4th ed. International Labour Organization, 1998.
- Sundquist, A., Bigdeli, S., Jalili, R., Druzin, M. L., Waller, S., Pullen, K. M., El-Sayed, Y. Y., Taslimi, M. M., Batzoglou, S. and Ronaghi, M. Bacterial flora-typing with targeted, chip-based Pyrosequencing. BMC microbiology, 2007; 7: 108.
- Suzuki, H., Ohnishi, Y. and Horinouchi, S. Arylamine N-acetyltransferase responsible for acetylation of 2-aminophenols in Streptomyces griseus. Journal of bacteriology, 2007; 189: 2155-2159.
- Takeo, M., Fujii, T. and Maeda, Y. Sequence analysis of the genes encoding a multicomponent dioxygenase involved in oxidation of aniline and o-toluidine in Acinetobacter sp. strain YAA. Journal of fermentation and bioengineering, 1998a; 85: 17-24.
- Takeo, M., Fujii, T., Takenaka, K. and Maeda, Y. Cloning and sequencing of a gene cluster for the meta-cleavage pathway of aniline degradation in Acinetobacter sp. strain YAA. Journal of fermentation and bioengineering, 1998b; 85: 514-517.
- Takeo, M., Ohara, A., Sakae, S., Okamoto, Y., Kitamura, C., Kato, D. and Negoro, S. Function of a glutamine synthetase-like protein in bacterial aniline oxidation via ã-glutamylanilide. Journal of bacteriology, 2013; 195: 4406-44014.
- Tanaka, T., Hachiyanagi, H., Yamamoto, N., Iijima, T., Kido, Y., Uyeda, M. and Takahama, K. Biodegradation of endocrine-disrupting chemical aniline by microorganisms. Journal of health science, 2009; 55: 625-630.
- Tuah, P. M., Jamaluddin, H., Hamzah, S. H., Haron, R., Yahya, A., Salleh, M. M. and Rashid, N. A. 2004. Biodegradation of phenol by locally isolated strains from petrochemical wastewater treatment plants. Environmental Biotechnology: Advancement in Water and Wastewater Applications in the Tropics: Selected Proceedings of the IWA International Conference on Environmental Biotechnology, Held in Kuala Lumpur, Malaysia, 9-10 December 2003. IWA Publishing, p. 109.
- Urata, M., Uchida, E., Nojiri, H., Omori, T., Obo, R., Miyaura, N. and Ouchiyama, N. Genes involved in aniline degradation by Delftia acidovorans strain 7N and its distribution in the natural environment. Bioscience, biotechnology, and biochemistry, 2004; 68: 2457-2465.
- Vangnai, A. S. and Petchkroh, W. Biodegradation of 4-chloroaniline by bacteria enriched from soil. FEMS microbiology letters, 2007; 268: 209-216.
- Wang, J. Z., Guan, Y. F., Ni, H. G., Luo, X. L. and Zeng, E.Y. Polycyclic aromatic hydrocarbons in riverine runoff of the Pearl River Delta (China): concentrations, fluxes, and fate. Environmental science & technology, 2007; 41: 5614-5619.
- Wang, Y. J., Zheng, Y. G., Xue, J. P. and Shen, Y. C. Microbial transformation of indole-3-acetonitrile to indole-3-acetamide by Nocardia sp. 108. Process Biochemistry 41, 1746-1750.
- Wyndham, R.C., (1986) Evolved aniline catabolism in Acinetobacter calcoaceticus during continuous culture of river water. Applied and Environmental Microbiology, 2006; 51: 781-789.
- Zeyer, J., Wasserfallen, A. and Timmis, K. N. Microbial mineralization of ring-substituted anilines through an ortho-cleavage pathway. Applied and Environmental Microbiology, 1985; 50: 447-453
- Zhang, T., Zhang, J., Shuangjiang, L. and Zhipei, L. A novel and complete gene cluster involved in the degradation of aniline by Delftia sp. AN3. Journal of Environmental Sciences, 2008; 20: 717-724.
- Zhuang, R., Zhong, W., Yao, J., Chen, H., Tian, L., Zhou, Y., Wang, F., Bramanti, E. and Zaray, G. Isolation and characterization of aniline-degrading Rhodococcus sp. strain AN5. Journal of Environmental Science and Health Part A, 2007; 42: 2009-2016.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.