ISSN: 0973-7510
E-ISSN: 2581-690X
Pleurotus sp is an edible mushroom, which contains essential nutrients and medicinal properties for living thing. Molecular characterization and genome differentiation of pleurotus sp. studied using gel electrophoresis, RAPD and PCR techniques. Separated DNA was amplified using internal transcribed spacer primers. 5.8s rRNA sequence submitted to Genbank for comparison and to obtain accession number. Differentiation and identification of Pleurotus sp found by similarity searches (Accession number: JQ740170 and JQ740171). Comparative studies have shown these two Pleurotus sp (spp1 & ssp2) are closely related to species, 95% – 100% identical, Pleurotus Citrinopileatus, Pleurotus populinus, Pleurotus ostreatus, Pleurotus abieticola, Pleurotus pulmonarius, Pleurotus eryngii and Pleurotus sapidus. These Pleurotus sp have been studied for exhibiting pharmacological potential activities such as antioxidant, antitumor, immunomodulatory activity and hypocholesterolemic through sequence similarity, nucleotide composition, evolutionary studies, secondary structure prediction and conserved region. The study extended to identify the significance of species diversity and fungal samples among various applications. Pleurotus (sp) sample was subjected to identification of bioactive compounds by using GC-MS. In Pleurotus spp1: Pyridine- 3-carboxamide, 4-dimethylamino-N-(2, 4-difluorophenyl), Piperidin-4-carboxylic acid, Aspidofractinine-3-methanol, (2à, 3á, 5à), Indolizine, and 2-(4-methylphenyl)-. Pleurotus spp2: Imidazolidine, 1, 3-dinitro, Phenol, 2-methyl-4-(1, 1, 3, 3-tetramethylbutyl), and Squalene. Enhancement of more entries as well as sequence comparison and virtual screening might help researchers to acknowledge real world problems between cultivating nutritional, medicinal and poisonous mushroom.
Edible Mushroom, internal transcribed spacer, metabolites, pharmacological activities, species diversity
Mushrooms are having highly nutritive and medicinal values so now a day’s peoples are readily used to cultivate in controlled biological environment and it has been extensively used as food since ancient time (Sathyaprabha et al. 2011). Although, mushrooms are rich in proteins and fibers it contain low amount of calories and fat, in the presence of low fat, peoples are highly used the edible mushroom frequently (Manzi et al., 2001). Cultivation of Pleurotus spp1 and spp2 in different substrates. Spawn were prepared in three types of substrates such as sorghum grain, Paddy grain and Maize grain. For fruit body cultivation Paddy straw, Sugar cane trash, Teak Leaves, Black gram Pods, and Banana Leaves are the substrates. Paddy grains substrate was good in growing and gives high yield of mycelium when compared to the sorghum grains and maize grain (Sathyaprabha et al., 2011).
In the presence of good nutritive value of mushrooms, a remarkable progress required in the breeding of its new strains which results in difficulty of their identification. Need of advance techniques for species identification beyond morphological and physiological criteria, because these characteristics highly influenced by cultivating conditions were posed big challenge (Staniaszek et al., 2002; Iqbal et al., 2010) . The genetic diversity of mushrooms has been worked out using molecular markers especially random amplified polymorphic DNA (Staniaszek et al., 2002; Stajic et al., 2005; Ravash et al., 2009). RAPD is used to assess genetic similarity and phylogenetic analysis due to the simplicity in its methodology and utility (Gepts 1993).
(Ro et al., 2007) studied the diversity of Pleurotus eryngii using RAPD technique and its sequence analysis and observed that, grouping based on physiological parameters is closely related to RAPD based grouping. (Gepts, 1993) used randomly amplified polymorphic DNA technique to access the genetic diversity among 37 pleurotus species of mushrooms. In another study, polymerase chain reaction (PCR) amplification was used to evaluate the genetic diversity among 45 pleurotus strains and found that, this technique was better than morphological analysis (Rajaratnam and Thiagarajan 2012). Identified and compared three medicinal mushroom (Ganoderma lucidum, Coriolus versicolor, and Fomes fomentarius) species from Korea based on nuclear large subunit rDNA sequences (Dung et al., 2012). Nucleotide sequencing of Termitomyces albuminosus (Lee et al., 2006; Yang et al., 2012) and Agaricus bisporus (Chen et al., 2011) among others was also performed and deposited into sequence database. Available sequence entry and information in genbank database for mushroom is not sufficient (Morin et al., 2012). A primer based on ITS region is rather useful for molecular characterization in fungi at the species level and within the species (Sudip Kumar Das et al., 2013; Williams et al., 1990; Chen et al., 2001). Keeping in view the usefulness of morphological and molecular markers, the present study was planned to investigate the genetic diversity among different strains of cultivated mushroom (Pleurotus sp.). Fragments of the DNA were amplified using ITS1 (Internal Transcribed Spacers) and ITS4 primers. The nucleotide sequences of mushrooms were matched from the available known sequences of genbank database.
Today, with the advancement of techniques and nutrient values mushrooms have occupied an important place in food in several parts of the world. We found that mushrooms are healthy foods, even though they are deficient in calories and fat and consist of about 90% water (Sathyaprabha et al., 2011). Mushrooms have been reported to be of therapeutic value, useful in preventing diseases such as hypertension, Hypercholesterolemia, cancer and also having antibacterial and antiviral properties. Functional characteristics are mainly due to their chemical composition. Pleurotus sp were further studied for screening bioactive compounds and vitamins using GC-MS. (Suresh Lalitharani 2010; Sathyaprabha et al., 2011).
DNA extraction and isolation from mushroom fruit body
0.01 g of mycelium was grinded with mini grinder using 75 µl of STE extraction buffer (320 mM Sucrose, 10 mM Tris-Cl, 20 mM EDTA, 75 mM NaCl and 2.5 ml of 20% SDS) along with 5 mg of Polyvinyl pyrolidone and 0.1 g of silica powder, incubated at 65°C for 10 minutes. Centrifuged the sample at 13,000 rpm for 10 minutes. To the supernatant, equal volume of chloroform: isoamyl alcohol was added and repeated the centrifugation. To the aqueous layer, added 2/3 volume of isopropanol and centrifuged at 13,000 rpm for 10 min. The pellet was washed with 70% ethanol by centrifuging and the pellet was dried, dissolved in 50µl TE buffer.
The DNA stock samples was quantified using UV spectrophotometer at 260 and 280 nm using the convention that one absorbance unit at 260 nm wavelength equals 50 µg DNA per ml. The Ultra Violet (UV) absorbance was checked at 260 and 280 nm for determination of DNA concentration and purity. Purity of DNA was judged by the optical density ratio at 260:280 nm. The DNA having ratio between1.8 – 2.0 was considered to be of good purity. Concentration of DNA was estimated using the formula. Concentration of DNA (mg/ml) = OD 260 x 50 x Dilution factor Quality of DNA was again checked by Agarose gel electrophoresis. The 0.8% Agarose was prepared (0.8 g Agarose power / 100 ml 1 X TBE), and was melted. 30 ml Agarose was poured into the casting tray. The gel was allowed to solidify and the comb and tape was removed. 1 X TBE (Tris-Borate-EDTA; electrophoresis buffer) was added to the chamber until the buffer just covers the top of the gel. The samples were loaded with Bromophenol blue loading dye, taking care not to puncture the well bottoms. The power pack was turned on and run at 100 V. The gel was viewed on a UV transilluminator after electrophoresis.
DNA Amplification
The DNA was used further for PCR amplification. ITS (Internal Transcribed Spacer) fragment was amplified by PCR from fungal genomic DNA using ITS-Primer “ITS 1: 5 ’-TCCGTAGGTGAACCTGCGG-3’ “,” ITS4: 5’- TCCTCCGCTTATTGATATGC-3’ “. PCR was carried out in a final reaction volume of 25 µl in 200 µl capacity thin wall PCR tube. Composition of reaction mixture for PCR is given in PCR tubes containing the mixture were tapped gently and spin briefly at 10,000 rpm. The PCR tubes with all the components were transferred to thermal cycler. The PCR protocol designed for 30 cycles for the primers used is given below. Deionized water 1 6.5 µl, Taq buffer without MgCl2 (10 X) 2.5µl, MgCl2 (15 mM) 1.0 µl, Forward Primer (10 pm/µl) 2.0 µl, Reverse Primer (10 pm/µl) 0.5 µl,Taq DNA Polymerase (5U/µl) 0.5 µl, Template DNA (20 ng/µl) 0.5 µl, Final Volume 25.0 µl. Initial Denaturation 95°C for 5 minutes, Denaturation 94°C for 30 seconds, annealing 55°C for 30 seconds, extension 72°C for 45 seconds, and final elongation 72°C for 10 minutes end.
Analysis of DNA Amplification by Agarose Gel Electrophoresis
Loaded 5µl of PCR product with 4 µl bromophenol blue (Loading Dye) in 1.5% Agarose gel and ran the gel at constant voltage of 100V and current of 45ºA for a period of 1 hr 20 min till the bromophenol blue has travelled 6 cms from the wells. Viewed the gels on UV transilluminator and photograph of the gel was taken (Sudip Kumar Das et al., 2013).
DNA Sequencing
Amplified PCR product was purified using column purification as per manufacturers Guidelines, and further used for sequencing. The concentration of the purified DNA was determined and subjected to automated DNA sequencing on ABI3730 XL Genetic Analyzer (Applied Bios stems, USA).
ITS Sequence alignment and analysis
Each nucleic acid sequence was edited manually trimmed to remove unreadable sequence at the 3 ’and 5’ ends (considering peak and Quality Values for each base) using the sequence analysis tools. The edited sequences were then used for similarity searches using BLAST (Basic Local Alignment Search Tool) programmed in the NCBI Genbank (www.ncbi.nlm.nih.gov/blastn) database for identifying the fungal strains (Accession number: JQ740170 and JQ740171) (Senthil et al. 2011).
Qualitative analysis for phytochemical constituents on Pleurotus spp1 and Pleurotus spp2
Aqueous extract of Pleurotus spp1 and Pleurotus spp2 samples were used to carry out the Qualitative analysis using standard procedures to identify the phytoconstituent as described (Sofowora 1993). Test for tannins: 0.5 gm of the fresh mushroom samples were taken in test tubes boiled with 20 ml of water and filtered. A few drops of 0.1% ferric chloride was added and observed for brownish green or a blue-black coloration. Test for phlobatannins: Deposition of a red precipitate, when an aqueous extract of each plant sample was boiled with 1% aqueous hydrochloric acid was taken as evidence for the presence of Phlobatannins. Test for saponin: About 2 gm of the fresh sample was boiled in 20 ml of distilled water in a water bath and filtered. 10 ml of the filtrate was mixed with 5 ml of distilled water and shaken vigorously for a stable persistent froth. The frothing was mixed with 3 drops of olive oil and shaken vigorously, then observed for the formation of emulsion. Test for flavonoids: 5ml of the diluted ammonia solution was added to a portion of aqueous filtrate of plant extract followed by the addition of concentrated sulphuric acid shown formation of yellow color. Test for steroids: 2 ml of acetic anhydride was added to 0.5 g ethanol extract of each sample with 2 ml H2S04. The color changed from violet to blue or green in some samples indicating the presence of steroids. Test for terpenoids: 5 ml of each extract was mixed in 2 ml of chloroform, and concentrated H2S04 (3 ml) was carefully added to form a layer. A reddish brown coloration of the interface was formed to show positive results for the presence of Terpenoids. Test for cardiac glycosides: 2 ml of glacial acetic acid containing one drop of ferric chloride solution was added to 5ml of the mushroom extract. Addition of 1ml concentrated sulphuric acid shown formation of a brown ring at the interface indicates the presence of cardiac glycosides.
Identification Bioactive compounds by GC-MS – Perkin Elmer
Sample preparation
Twenty five gram of smashed fresh Pleurotus spp1 and spp2 sample was taken in a conical flask with 30 ml of distilled ethanol and methanol crushed with respective solvent and stored it for overnight soaking. Then filter the sample and concentrated the sample with help of nitrogen flushing. Filter the filtrate with sodium sulphate. 3µl of purely prepared sample was injected into the programme GC-MS instrument.
Gas Chromatography Programme
Column: Elite-5MS (5% Diphenyl / 95% Dimethyl poly siloxane), 30 x 0.25mm x 0.25mm df Equipment: GC Clarus 500 Perkin Elmer, Carrier gas: 1ml per min, Split: 10:1, Detector: Mass detector Turbo mass gold-Perkin Elmer, Software: Turbomass 5.2, Sample injected: 3ml. Oven temperature Programme: 110° C -2 min hold ,Up to 200°C at the rate of 10° C/min-No hold, Up to 280°C at the rate of 5°C / min-9 min hold ,Injector temperature 250°C, Total GC running time 36 min. Mass Spectrum Programme: Library used NIST Version-Year 2005, Inlet line temperature 200°C, Source temperature 200°C Electron energy: 70 eV, Mass scan (m/z): 45-450, Solvent Delay: 0-2 min, Total MS running time: 36 min.
Virtual screening
Till date, virtual screening of compound aids to identify their interaction with proteins. Protein-ligand interactions found by Insilico analysis will exhibit the potential of bioactive compounds and to obtain lead compounds rather than chemically synthesis. Out of 17, which are known as antitumor and anticancer agents through existing methods, piperidin-4-carboxylic acid and squalene, were taken (Fig. 4 & 5). Phenol used to treat and aid for microbial and fungal infection was shown in fig. 6. It is noteworthy that screening all metabolites on various medicinal properties.
Genomic DNA
Genomic DNA was extracted from harvested fruit body of Pleurotus sp (SPP1 & SPP2). Extracted DNA molecules were performed by Agarose Gel electrophoresis techniques. Genomic DNA was isolated from the fungal sample/culture provided. Its quality was evaluated on 0.8% Agarose Gel and a single band of high-molecular weight DNA was observed. ITS fragment was amplified by PCR from the above isolated genomic DNA. The PCR amplicon was purified by column purification in order to remove contaminants. Commercially available 100 bp ladder was used as standard molecular weight DNA. A single discrete band was observed when resolved on Agarose Gel (Fig. 1). Extracted DNA from the fruit body of Pleurotus sp mushroom subjected to nucleotide sequencing using ITS1 and ITS4 conserved primer stretches. The sequence was aligned using similarity searches algorithm and aligned sequence (~800 bp) revealed 95% – 100% matched score with Pleurotus sp (Fig. 2). Characterized Pleurotus spp1 (https://www.ncbi.nlm.nih.gov/nuccore/JQ740170) and Pleurotus spp2 (https://www.ncbi.nlm.nih.gov/nuccore/JQ740171) based on phenotype, genotype and molecular data found that both were unique to each other (Fig. 1 & 3). In addition GC content of Pleurotus sp ranged from 44 – 45% with a constant ratio (Table 1). Subsequently, Pleurotus sp identical were retrieved for the phylogenetic analysis. Nine species grouped together one clad, pointing their diversification and resemblance (Fig. 3). A phylogenetic analysis- maximum-likelihood based on 5.8s rRNA was conducted to determine the relationship of Pleurotus sp with other medically important species. Except, sequence data of P. sapidus due to its long order in length.
Table (1):
Composition and conserved region found among Pleurotus sp.
S. No |
Name of the organism |
Accession No |
Isolate |
Bp |
GC % |
AT % |
|---|---|---|---|---|---|---|
1 |
Pleurotus spp1 |
JQ740170 |
Spp1 |
786 |
44.53 |
55.47 |
2 |
Pleurotus spp2 |
JQ740171 |
Spp2 |
774 |
44.44 |
55.06 |
3 |
Pleurotus citrinopileatus |
JF736661 |
NE01 |
782 |
44.88 |
55.12 |
4 |
Pleurotus populinus |
AYA450346 |
9936 |
781 |
44.94 |
55.06 |
5 |
Pleurotus ostreatus |
AY450345 |
6689 |
782 |
44.88 |
55.12 |
6 |
Pleurotus abieticola |
AY450348 |
6554 |
780 |
44.49 |
55.51 |
7 |
Pleurotus pulmonarius |
AY450349 |
4203 |
773 |
44.50 |
55.50 |
8 |
Pleurotus eryngii |
AY450347 |
6690 |
781 |
45.20 |
54.80 |
9 |
Pleurotus sapidus |
JF736664 |
NE07 |
1123 |
44.97 |
55.03 |
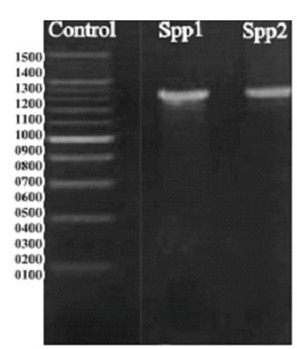
Fig. 1. Amplification of Pleurotus sp with ITS primers and separation of the PCR products by gel electrophoresis (spotted ~ 600-800 bp): Lane 1. DNA marker; Lane 2 and 3: Pleurotus spp1&spp2. Gel electrophoresis marker ranges bands from 1500 bp to 100 bp while sample shows that approximately at 1200 bp to 1100 bp.
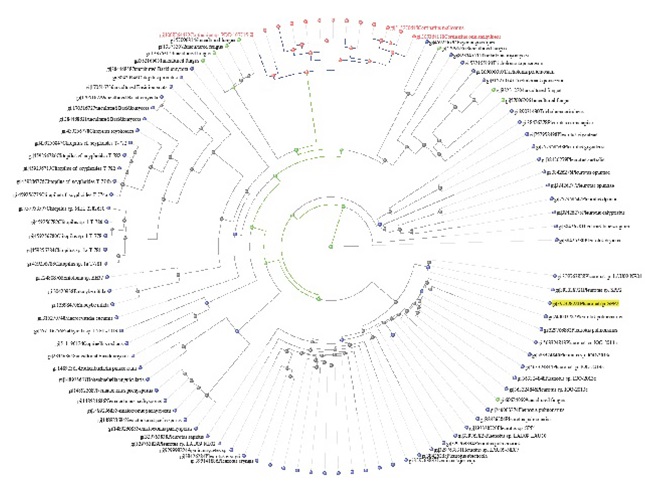
Fig. 2. Circular Tree View of Pleurotus sp based on Jukes-Cantor Correlated Distance model (0.004) including other species.
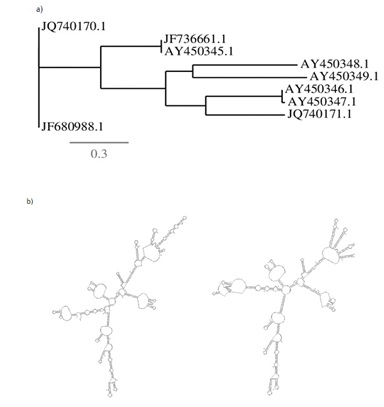
Fig. 3. a) Tree based on the rRNA-ITS sequence using maximum-likelihood phyloge-nies, shown relationships among Pleurotus sp (JQ740170 and JQ740171) and b) secondary structure predicted using mFOLD server for their identity (spp1 & spp2).
Phylogenetic tree constructed shown a close relationship between Pleurotus Citrinopileatus and Pleurotus ostreatus, Pleurotus abieticola and Pleurotus pulmonarius. Pleurotus populinus and Pleurotus eryngii with Pleurotus spp1 & spp2, grouped together for evolutionary studies. The significance of the work is to gain proper identification knowledge of the edible and non-edible mushrooms, which may drive towards domestication and commercialization of the wild species for economic benefits, nutritional value and medically important fungi instead deriving molecular taxonomy (Table 2).
Table (2):
5.8s rRNA-ITS sequences/species of closely related to Pleurotus spp1 and spp2 studied for Phytoconstituent and medicinal values (Chen et al. 2011; Jose et al. 2002; Bobek and Ozdin 1996; Fu et al. 2009; Selegean et al. 2009; Daba and Ezeronye 2003; Adebayo et al. 2012)
S. No |
Name of the organism |
Identity (%) |
Accession No |
Studied and found Medicinal Properties |
|---|---|---|---|---|
1. |
Pleurotus Citrinopileatus |
98 |
JF736661 |
Anti inflammatory, Anti oxidant, and Anti tumor activities |
2. |
Pleurotus ostreatus |
98 |
AY450345 |
Anti tumor activity, Immunomodulator activity, Anti oxidant activity, Hypocholesterolemic |
3. |
Pleurotus abieticola |
98 |
AY450348 |
Anti inflammatory activity, Anti tumor activities and Anti oxidant, |
4. |
Pleurotus populinus |
97 |
AYA450346 |
Prostate cancer and other cancers. |
5. |
Pleurotus pulmonarius |
97 |
AY450349 |
Anti inflammatory, Anti oxidant and Anti tumor activities |
6. |
Pleurotus eryngii |
97 |
AY450347 |
Anti inflammatory |
7. |
Pleurotus sapidus |
96 |
JF736664 |
Anti viral, Anti bacterial, Anti diabetic, Anti arthritic, Anti tumor activities |
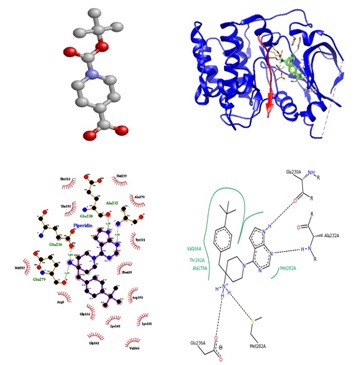
Fig. 4. Structure of 4-(4-tert-butylbenzyl)-1-(7h-pyrrolo (2, 3-d) pyrimidin-4-yl) piperi-din-4-amine bound to protein kinase b (2XH5). Ball and stick representation of ligand (piperidin) shown with hydrogen donor and acceptor. Complex structure of receptor and ligand was shown in rib-bon model. Schematic diagram of bonded and non bonded interaction was shown in yellow and black dot line (ligplot and pose view).
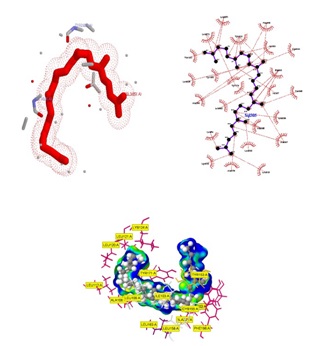
Fig. 5. Crystal structure of supernatant protein factor bound to squalene (PDB: 4OMK). Ball and stick representation of ligand (squalene) shown with active site residues
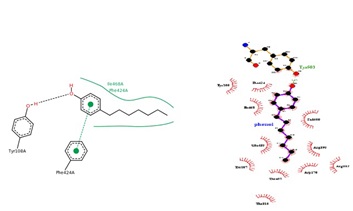
Fig. 6. Structure of the flavoenzyme vanillyl-alcohol oxidase in complex with phenol was shown (PDB: 1AHZ). Schematic representation of phenol with receptor has shown for the affinity and enzyme inhibition (Pose view and ligplot).
Qualitative analysis
Qualitative analysis of Phytochemical revealed that, Pleurotus spp1 shows the high concentration of tannin when compared to sugarcane trash and teak leaves, phlobatannins was present in paddy straw but absent in all other four substrates. Saponin were absent in paddy straw but present in all other four substrate. Flavonoid was present in all substrates steroids was shows that high concentration in all the substrates. Terpenoids were absent in paddy substrates and banana leaves substrates, but present in sugarcane trash, teak leaves and black gram pods. Cardiac glycosides were present in all the substrates while absent in black gram pods (Table 3).
Table (3):
Pharmacological properties found in Pleurotus spp1 and spp2
| S. No | Parameters | Pleurotus spp1 | Pleurotus spp2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S4 | S5 | S1 | S2 | S3 | S4 | S5 | ||
| 1. | Tannin | +++ | ++ | ++ | +++ | +++ | —– | +++ | +++ | —– | +++ |
| 2. | Phlobatannins | +++ | —- | — | —- | —- | —– | +++ | ——- | —— | —- |
| 3. | Saponin | —– | ++ | +++ | ++ | ++ | +++ | —– | +++ | ++ | ++ |
| 4. | Flavonoids | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 5. | Steroids | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 6. | Terpenoids | —– | +++ | ++ | ++ | —- | —– | —– | +++ | ++ | —– |
| 7. | Cardiac glycosides | +++ | +++ | +++ | —- | +++ | +++ | —– | +++ | —— | +++ |
Note: Present – +++: Slightly Present – ++: Absent – —–
S1: Paddy straw, S2: Sugarcane trash, S3: Teak leaves, S4: Black gram pods, S5: Banana leaves
In Pleurotus spp2 Tannin, phlobatannins and Terpenoids were absent in paddy straw substrates but shows the presence of saponin, flavonoids, steroids but absences of saponin, Terpenoids and cardiac glycosides. Phlobatannins were present in the sugarcane trash substrates but absent in all other substrates, paddy straw, teak leaves, black gram pods and banana leaves. Black gram pods show the absence of tannin, Phlobatannins and cardiac glycosides, while flavonoids and steroids were strongly present but presence of saponin and terpenoids. Terpenoids and phlobatannins were absent in banana leaves but tannin, flavonoids, steroids and cardiac glycosides were strongly present while saponin present (Table 3).
Identification of Bioactive Compounds in Phytochemical properties found by qualitative analysis suggesting their roles in synthesis of bioactive compound and vitamins. According to the results, the Phytocomponents screened were shown in the (Table 4 and 5). Animals and Human might require vitamins for their routine activities, in addition to food. Dissemination of vitamins has made with its dissolving nature and storage system. (Sathyaprabha et al., 2011).
Table (4):
Bioactive Components identified in the ethanol extract of Pleurotus spp1
No. |
RT |
Name of the compound |
Molecular Formula |
MW |
Peak Area % |
Group |
|---|---|---|---|---|---|---|
1. |
7.17 |
DL-Alanine, N-benzoyl-N-(3-chloro-4-fluorophenyl)-, methyl ester |
C17H15ClFNO3 |
335 |
0.76 |
Fatty Acid |
2. |
7.44 |
(1H)Pyrrole-2-carboxaldehyde, 4-(trichloroacetyl)- |
C7H4Cl3NO2 |
239 |
0.76 |
Aromatic organic compounds |
3. |
7.96 |
2-Amino-4-hydroxy-6,7,8-trimethylpteridine |
C9H11N5O |
205 |
3.80 |
Heterocyclic |
4. |
12.49 |
Benzoic acid 1-methoxy-1H-tetrazol-5-ylmethyl ester |
C10H10N4O3 |
234 |
0.76 |
Fatty acid |
5. |
13.06 |
Pyridine-3-carboxamide, 4-dimethylamino-N-(2,4-difluorophenyl)- |
C14H13F2N3O |
277 |
74.14 |
Heterocyclic |
6. |
13.45 |
à-Ethyl aspartate |
C6H11NO4 |
161 |
2.28 |
Esters |
7. |
18.80 |
Piperidin-4-carboxylic acid |
C6H11NO2 |
129 |
1.52 |
Heterocyclic |
8. |
20.82 |
Aspidofractinine-3-methanol, (2à,3á,5à)- |
C20H26N2O |
310 |
4.94 |
Alkaloids |
9. |
24.67 |
Indolizine, 2-(4-methylphenyl)- |
C15H13N |
207 |
11.03 |
Alkaloids |
Table (5):
Bioactive Components identified in the ethanol extract of Pleurotus spp2
No. |
RT |
Name of the compound |
Molecular Formula |
MW |
Peak Area % |
Group |
|---|---|---|---|---|---|---|
1. |
7.96 |
5-(4-Hexyloxybenzoyloxy)-2-(4-nitrophenyl) pyrimidine |
C23H23N3O5 |
421 |
0.01 |
Heterocyclic |
2. |
10.15 |
Imidazolidine, 1,3-dinitro- |
C3H6N4O4 |
162 |
0.02 |
Heterocyclic |
3. |
12.64 |
dl-Alanine |
C3H7NO2 |
89 |
0.01 |
α-amino acid |
4. |
16.89 |
Phenol, 2-methyl-4-(1,1,3,3-tetramethylbutyl)- |
C15H24O |
220 |
0.07 |
Phenols |
5. |
17.51 |
1H-Tetrazol-5-amine |
CH3N5 |
85 |
0.02 |
organic compound |
6. |
20.90 |
Aspidofractinine-3-methanol, (2à,3á,5à)- |
C20H26N2O |
310 |
99.83 |
Alkaloids |
7. |
24.66 |
Squalene |
C30H50 |
410 |
0.05 |
Triterpene |
Pleurotus sp was described as edible mushroom but the significance of species diveristy and nutrients , clinical values studied through amplification and ITS region sequence comparison. The ITS fragment of the genomic DNA of two edible mushrooms, collected from TNAU, Aduthurai, Thanjavur, India, was amplified using ITS1 and ITS4 primers and subjected to nucleotide sequence determination for identification of species. The amplification and sequence comparison uncovers species diversity and other species with similar molecular information for its domestication, characterization of nutritional value and medicinal benefits. Qualitative analysis made to estimate the pharmacological properties. By ranking strongly present, present and absent in 7 groups indicate that the availability and rich properties. Comparative study of the ranking method describes the abundance of flavonoids and steroids. The results obtained from GC-MS analysis is given in the (Table 4 and 5). Both pleurotus sp contains vitamins namely A, D3, E, K, niacin, pyridoxine, thiamine and riboflavin, were analyzed. Phyto constituents screened through GC-MS were studied for identification of compound nature and group. Chemical composition and secondary metabolites of pleurotus sp were studied. Totally seventeen metabolites screened were examined for different properties and grouped as those belonging to acids, alcohol, alkane, amides, esters, fatty acids, terpenoids, heterocyclic and phenols (Mohamed et al., 2014). Hetero cyclic compounds were appeared more in number than fatty acids, ester, aroma and phenols (Rivera et al., 2012).
Piperidin-4-carboxylic acid
Piperidin derivatives were found in GC-MS studies further investigated with virtual screening to access their potent as antitumor agent (McHardy et al., 2010). Here the complex structure demonstrated that how the piperidin derivatives bind to protein and inhibits. Interaction between the receptor and ligand studied based on substitution method ( McHardy et al., 2010) prepared piperidin moieties to verify it’s multifold with side chains (Fig.4).
Squalene
Squalene and squalene analogs studied by several techniques and found useful in cancer therapy. Still its availability and usage widens the research on the skin treatment (Huang et al., 2009) and cholesterol endosynthesis (Christen et al., 2015). Here supernatant protein factor (SPF) has modeled and studied for cholesterol endosynthesis (Fig.5).
Phenol
Phenolic compounds have known to possess medicinal values. (Alves et al., 2013) reported that phenolic compounds could be used for antimicrobial activity as activator and inhibitor. Especially for resistance bacteria, phenolic compounds had shown the high inhibition rate when compared to different concentration level. A Study and virtual screening carried out to negotiate the species diversity and medicinal properties among pleurotus sp. were available. Recent studies shown that heterocyclic and phenolic compounds, has more impact on cancer related studies.Sequence comparison and insilico analysis made it possible to explore molecules interaction with metabolites.
ACKNOWLEDGMENTS
We thank Dr. A. Panneerselvam, Professor, Department of Botany and Microbiology, A.V.V.M Sri Pushpam College, Thanjavur, TN- India for his valuable guidance and Tamil Nadu Agricultural University, Aduthurai, Thanjavur, TN-India for sample preparation.
- Adebayo EA, Oloke JK, Majolagbe ON, Ajani RA, Bora TC. Antimicrobial and anti-inflammatory potential of polysaccharide from Pleurotus pulmonarius LAU 09. Afri J Microbiol Res, 2012; 6:3315-3323.
- Alves MJ, Ferreira IC, Froufe HJ, Abreu RM, Martins A, Pintado M. Antimicrobial activity of phenolic compounds identified in wild mushrooms, SAR analysis and docking studies. Journal of applied microbiology. 2013; 115(2):346-57.
- Bobek P and Ozdin L, Oyster mushroom (Pleurotus ostreatus) reduces the production and secretion of very low density lipoproteins in hyper-cholesterolemic rates. Zeitschrift für Ernährungswissenschaft, 1996; 35: 249-252.
- Chen JN, de Mejia EG, Wu JS, Inhibitory Effect of a Glycoprotein Isolated from Golden Oyster Mushroom (Pleurotus citrinopileatus) on the lipopolysaccharide – induced inflammatory Reaction in RAW 264.7 Macrophage. J Agric Food Chem, 2011; 59:7092-7097.
- Chen YC, Eisner JD, Kattar MM, Rassoulian-Barrett SL, Lafe K, Bui U, Limaye AP, Cookson BT, Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. Journal of Clinical Microbiology, 2001; 39: 4042–4051.
- Chen S, Xu J, Liu C, Zhu Y, Nelson DR, Zhou S, Li C, Wang L, Guo X, Sun Y,Luo H, Li Y, Song J, HenrissatB, Levasseur A, Qian J, Li J, Luo X, Shi L, He L, Xiang L, Xu X, Niu Y, Li Q, Han MV, Yan H, Zhang J, Chen H, Lv A, Wang Z, Liu M, Schwartz DC, Sun C, Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat Commun, 2012; 26: 913.
- Christen M, Marcaida MJ, Lamprakis C, Aeschimann W, Vaithilingam J, Schneider P, Hilbert M, Schneider G, Cascella M, Stocker A, Structural insights on cholesterol endosynthesis: Binding of squalene and 2, 3-oxidosqualene to supernatant protein factor. Journal of structural biology. 2015; 190(3):261-70.
- Daba AS, Ezeronye OU, Anti-cancer effect of polysaccharides isolated from higher Basidiomycetes mushrooms. African Journal of Biotechnology, 2003; 2: 672-678.
- Das SK, Mandal A, Datta AK, Gupta S, Paul R, Saha A, Sengupta, S, Dubey PK, Nucleotide Sequencing and Identification of Some Wild Mushrooms. The Scientific World Journal, 2013; 2013: 403191.
- Dung NTP, Tuyen DB, Quang PH, Morphological and genetic characteristics of oyster mushrooms and conditions effecting on its spawn growing. International Food Research Journal, 2012; 19: 347–352.
- Fu HY, Shieh DE, HO CT, Antioxidant and free radical scavenging activities of edible mushrooms. Journal of Food Lipids, 2009; 9:35-43.
- Gepts P, The use of molecular and biochemical markers in crop Evolution studies. Evolutionary Biology, 1993; 27: 51-94.
- Huang ZR, Lin YK, Fang JY, Biological and pharmacological activities of squalene and related compounds: potential uses in cosmetic dermatology. Molecules. 2009; 14(1):540-54.
- Iqbal A, Sadia B, Khan AI, Awan FS, Kainth RA, Sadaqat HA, Biodiversity in the sorghum (Sorghum bicolor L. Moench) germplasm of Pakistan. Genetics and Molecular Research, 2009; 9: 756-764.
- Jose N, Ajith TA, Janardhanan KK, Antioxidant, anti inflammatory, and antitumor activities of culinary-medicinal mushroom Pleurotus pufmonanus (Fr.) Quel. (Agaricomycetideae). International Journal of Medicinal Mushrooms 2002; 4: 329-335.
- Lalitharani S, Mohan VR, Selial Regini G, GC-MS analysis of ethanolic extract of Zanthoxylum rhetsa (ROXB.) DC spines. Journal of Herbal Medicine and Toxicology, 2010; 4: 191-192.
- Lee JS, Lim MO, Cho KY, Cho JH, Chan;g SY, Nam DH, Identification of medicinal mushroom species based on nuclear large subunit rDNA sequences. Journal of Microbiology, 2006; 44: 29–34.
- Manzi P, Aguzzi A, Pizzoferrato L, Nutritional value of mushrooms widely consumed in Italy. Food Chemistry, 2001; 73:321-325.
- McHardy T, Caldwell JJ, Cheung KM, Hunter LJ, Taylor K, Rowlands M, Ruddle R, Henley A, de Haven Brandon A, Valenti M, Davies TG, Discovery of 4-amino-1-(7 H-pyrrolo [2, 3-d] pyrimidin-4-yl) piperidine-4-carboxamides as selective, orally active inhibitors of protein kinase B (Akt). Journal of medicinal chemistry 2010; 53:2239-49.
- Mohamed EM, Farghaly FA, Bioactive compounds of fresh and dried Pleurotus ostreatus mushroom. International journal of biotechnology for wellness industries. 2014; 3(1):4-14.
- Morin E, Kohler A, Baker AR, Foulongne-Oriol M, Lombard V, Nagye LG, Ohm RA., Patyshakuliyeva A, Brun A, Aerts AL, Bailey AM, Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proceedings of the National Academy of Sciences, 2012; 109: 17501–17506.
- Rajaratnam S, Thiagarajan T, Molecular characterization of wild mushroom. European Journal of Experimental Biology 2012; 2:369–373.
- Ravash R, Shiran B, Alavi A, Zarvagis J, Evaluation of genetic diversity in Oyster mushroom (Pleurotus eryngii) isolates using RAPD marker. Journal of Science ;and Technology of Agriculture and Natural Resources, 2009; 13: 729-739.
- Rivera A, Quiroga D, Rios-Motta J, Dusek M, Fejfarova K, 1, 3-Dinitrosoimidazolidine. Acta Crystallographica Section E: Structure Reports Online. 2012; 68(8):2440.
- Ro HS, Kim SS, San Ryu J, Jeon CO, Lee TS, Lee HS, Comparative studies on the diversity of the edible mushroom Pleurotus eryngii: ITS sequence analysis, RAPD fingerprinting, and Physiological characteristics. Mycological Research 2007; 111: 710-715.
- Sathyaprabha G, Kumaravel S, Panneerselvam A, Bioactive Compounds Identification of Pleurotus platypus and Pleurotus eous by GC-MS. Adv Appl Sci Res, 2011; 2: 51-54.
- Sathyaprabha G, Kumaravel S, Panneerselvam A, Studies on phytochemical and vitamin analysis of Pleurotus platypus and Pleurotus oeus by GC-MS AND HPLC technique. International Journal of Pharmaceutical Sciences and Research 2011; 2:2816-2821.
- Sathyaprabha G, Panneerselvam A, Kumaravel S, Cultivation of Pleurotus platypus and Pleurotus eous in different agricultural waste substrates. Journal of Pharmacy Research 2011; 4: 2543-2544.
- Selegean M, Putz MV, Rugea T, Effect of the polysaccharide extract from the edible mushroom Pleurotus ostreatus against infectious Bursal Disease Virus, International journal of molecular sciences 2009; 10: 3616-3634.
- Senthil R, Angel KJ, Malathi R, Venkatesan D, Isolation, identification and computational studies on Pseudomonas aeruginosa sp. strain MPC1 in tannery effluent. Bioinformation 2011; 6: 187-190.
- Sofowora A, Medicinal plants and Traditional medicine in Africa. Spectrum Books. Ltd. Ibadan, Nigeria 1993; 2: 270-289.
- Stajic M, Sikorski J, Wasser SP, Nevo E, Genetic similarity and taxonomic relationships within the genus Pleurotus (higher Basidiomycetes) determined by RAPD analysis. Mycotaxon 2005; 93: 247-255.
- Staniaszek M, Marczewski W, Szudyga K, Maszkiewicz J, Czaplicki A, Qian G, Genetic relationship between Polish and Chinese strains of the mushroom Agaricus bisporus (Lange) Sing., determined by the RAPD method. Journal of applied genetics 2002; 43: 43-48.
- Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV, DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic acids research, 1990; 18: 6531-6535.
- Yang F, Xu B, Zhao S, Li J, Yang Y, Tang X, Wang F, Peng M, Huang Z, De novo sequencing and analysis of the termite mushroom (Termitomyces albuminosus) transcriptome to discover putative genes involved in bioactive component biosynthesis. Journal of Bioscience and Bioengineering 2012; 114: 228–231.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


