ISSN: 0973-7510
E-ISSN: 2581-690X
Staphylococcus haemolyticus is a highly resistant opportunistic pathogen having close genomic relatedness with other virulent species of staphylococci. However, compared to Staphylococcus aureus and Staphylococcus epidermidis, little is known about the resistance genes of S. haemolyticus. The purpose of this study was to characterise antibiotic resistance genes in S. haemolyticus isolates. Standard microbiological techniques were used to identify and confirm 104 S. haemolyticus isolates included in the study. Antibiotic susceptibility testing and D-test were performed, followed by PCR amplification of various resistance determinants (mecA, ermA, ermC, msrA, aac(6′)-Ie-aph(2″), ant(4′)-Ia,aph(3′)-IIIa, tetK, tetM, dfrA, fusB, fusC, fusD and mupA). Methicillin resistance was observed in 93.3% of study isolates. The maximum number of isolates showed resistance to erythromycin (n=79, 76%), followed by ciprofloxacin (n=66, 63.5%) and cotrimoxazole (n=58, 55.8%). In the D-test, 8 isolates showed inducible (iMLSB) and 11 showed constitutive (cMLSB) resistance. Among the resistance determinants, mecA gene (93.3%) was the most prevalent, followed by dfrA (50.5%). Furthermore, aac(6’)-Ie-aph(2’’) and aph(3’)-IIIa combination was observed in 26.9% of isolates, and aac(6’)-Ie-aph(2’’) alone was present in 3.8% of isolates. Among the study isolates, 17.3% exhibited tetK gene, whereas only 1% exhibited tetM; a combination of tetK and tetM was observed in one isolate. The fusB and fusC were present in 11.5% of isolates, and 12.5% of the isolates were positive for mupA. In conclusion, the present study underlines the concern of increasing antibiotic resistance among S. haemolyticus isolates. Avoiding misuse/overuse of antibiotics along with continuous surveillance programs can reduce the spread of antibiotic resistance.
S. haemolyticus Resistance, Multidrug-resistant S. haemolyticus, Antibiotic Drug Resistance
Staphylococcus haemolyticus is an opportunistic pathogen and the second most frequently isolated coagulase-negative staphylococci (CoNS), with high degree of genetic relatedness to Staphylococcus aureus and Staphylococcus epidermidis.1 It has an average nucleotide sequence similarity of 75% with S. aureus and S. epidermidis. Thus, there is a high probability that S. haemolyticus could act as a reservoir of resistance genes and disseminate them, thereby posing a threat of antibiotic resistance in hospital setup.2 Another unique feature of S. haemolyticus genome is that it undergoes constant rearrangement due to the presence of various insertion sequences.3 Empirical treatment with broad-spectrum antibiotics and genomic diversity due to frequent genomic rearrangements have led to the selection of multi-resistant strains that slowly replace susceptible strains in hospitals. These findings are consistent with the fact that among CoNS, S. haemolyticus possesses the highest level of resistance to commonly used antibiotics.4,5 Despite being the second most commonly isolated CoNS, there is a paucity of available data regarding its antimicrobial resistance. Hence, this study aimed to elucidate the antibiotic pattern and molecular characterisation of resistance genes in S. haemolyticus isolates from various clinical samples.
Study Isolates
A total of 104 S. haemolyticus isolates were collected from a tertiary care centre in Chennai during March 2016-January 2017 and used for further research. Initial sampling and identification of CoNS using standard sampling methods were performed by technical experts from the Microbiology Laboratory of the tertiary care centre. Further identification and species confirmation were performed as described below. The sources of the collected isolates are given in Table 1.
Table (1):
Clinical sources of the study isolates.
Source of the isolate |
% (No. of isolates) |
|---|---|
Skin and Soft tissue infections |
51.9% (n=54) |
High vaginal swab |
25.9% (n= 27) |
Semen |
13.5% (n=14) |
Urine |
5.8% (n=6) |
Ascitic fluid |
1.9% (n=2) |
Sputum. |
1% (n=1) |
Identification and Confirmation of S. haemolyticus
Staphylococcus isolates were initially identified using standard microbiological techniques such as Gram staining, catalase test, oxidation-fermentation test, coagulase test (tube and slide coagulase), DNase test, and mannitol fermentation on mannitol salt agar. Further species confirmation was performed using alkaline phosphatase, ornithine decarboxylase, urease, novobiocin and polymyxin B susceptibility, and carbohydrate (maltose, mannose, trehalose and sucrose) fermentation tests.6
Phenotypic Screening of Antibiotic Resistance
(i) Antibiotic Susceptibility Testing
The Kirby-Bauer disc diffusion method was used to test antibiotic sensitivity using the following antibiotic discs at the concentrations mentioned: cefoxitin (30 µg), ciprofloxacin (5 µg), clindamycin (2 µg), trimethoprim-sulfamethoxazole (1.25/23.75 µg), erythromycin (15 µg), gentamicin (10 µg), linezolid (30 µg), tetracycline (30 µg), rifampicin (5 µg), fusidic acid (10 µg), mupirocin (200 µg), and vancomycin (30 µg).7 The zone diameter was measured and interpreted according to the Clinical Laboratory Standards Institute guidelines (CLSI, 2015).8 S. aureus ATCC 25923 was used as the control strain.
(ii) Detection of Methicillin Resistance
The Kirby-Bauer disc diffusion method was performed to screen for resistance to methicillin using cefoxitin antibiotic disc (30 µg).7 For S. haemolyticus, a zone diameter of ≤ 24 mm was considered methicillin resistant.
(iii) Detection of Inducible and Constitutive Clindamycin Resistance
The D-test was performed to detect inducible clindamycin resistance of the isolates, and the results were interpreted according to the CLSI guidelines, 2015.8 Briefly, erythromycin (15 μg) disc and clindamycin (2 μg) disc were placed 15 mm apart (measured from the edge of the disc) in a previously swabbed lawn culture of the isolates with growth matching the turbidity of 0.5 McFarland standard. The zone of inhibition was observed the following day after incubation at 37°C. Blunting of the clindamycin zone near the erythromycin antibiotic disc (D-shape) showed inducible macrolide-lincosamide-streptogramin B (iMLSB) resistance phenotype, whereas resistance to both erythromycin and clindamycin showed constitutive resistance (cMLSB).
Genotypic Methods
DNA Extraction and Polymerase Chain Reaction
DNA extraction from all the study isolates was performed using the boiling lysis method; the extracted DNA was amplified for each of the resistance genes by polymerase chain reaction (PCR) in Mastercycler® Gradient (Eppendorf, Hamburg, Germany). The PCR products were then subjected to agarose gel electrophoresis, and the respective bands were visualised using Gel Logic 212 PRO imaging system. Analysis was carried out using the Carestream Molecular Imaging Software (Carestream Health, Incorporated, USA).
(i) Molecular Confirmation of S. haemolyticus
PCR amplification of the mvaA gene was performed for molecular confirmation of S. haemolyticus isolates.9
(ii) Genes conferring Antibiotic Resistance
The genes conferring resistance screened in the study were as follows: mecA– gene conferring methicillin resistance, aac(6’)-Ie-aph(2’’), aph(3’)-IIIa and ant(4′)– aminoglycoside modifying enzymes, msrA, ermA and ermC– genes conferring macrolide resistance, dfrA– gene conferring trimethoprim resistance, tetK and tetM– genes conferring tetracycline resistance, fusB, fusC and fusD– fusidic acid resistant genes, mupA– mupirocin resistant gene. The primers, PCR cycling conditions, and reference for the respective resistance determinants are shown in Table 2.
Table (2):
Primer sequence, product size, PCR cycling conditions and reference of the various genes screened in the study.
| Gene | Sequence | Product size (bp-basepair) | Cycling condition (˚C-degree Celsius, min-minute, sec-seconds) | Reference | ||
|---|---|---|---|---|---|---|
| Denaturation | Cycles | Final extension | ||||
| mvaA | F:5’GGTCGCTTAGTCGGAACAAT-3’ | 271bp | 92˚C for 3 min | 30 cycles of 92˚C -1 min, 56˚C -1 min, 72 ˚C -1min | 72 ˚C for 3 min | 9 |
| F:5’CACGAGCAATCTCATCACCT-3’ | ||||||
| mecA
|
F:5’TGCTATCCACCCTCAAACAGG-3’ | 286bp | 94˚C for 4min | 25 cycles of 94˚C-30sec, 54˚C-30sec, 72 ˚C -1min | 72 ˚C for 5 min | 10 |
| R:5’AACGTTGTAACCACCCCAAGA-3’ | ||||||
| aph(3)-III | F:5’CGATGTGGATTGCGAAAACT-3’ | 175 bp | 94˚C for 4min | 30 cycles of 94˚C-1 min, 57˚C-2 min, 72 ˚C- 1min | 72 ˚C for 5 min |
11 |
| R:5’CACCGAAATAACTAGAACCC-3’ | ||||||
| aac(6)-aph(2) | F:5’CATTATACAGAGCCTTGGGA-3’ | 279 bp | ||||
| R:5’AGGTTCTCGTTATTCCCGTA-3’ | ||||||
| ant(4)-I | F:5’ATGGCTCTCTTGGTCGTCAG-3’ | 367 bp | ||||
| R:5’TAAGCACACGTTCCTGGCTG-3’ | ||||||
| ermA | F:5’AAGCGGTAAACCCCTCTGA -3’ | 190bp | 94˚C for 4min | 30 cycles of 94˚C-1 min, 54˚C-30 sec, 72 ˚C-1min | 72 ˚C for 5 min |
12 |
| R:5’TTCGCAAATCCCTTCTCAAC-3’ | ||||||
| ermC | F:5’AATCGTCAATTCCTGCATGT-3’ | 299bp | ||||
| R:5’TAATCGTGGAATACGGGTTTG-3’ | ||||||
| tetK | F:5’GTAGCGACAATAGGTAATAGT-3’ | 360bp | ||||
| R:5’GTAGTGACAATAAACCTCCTA-3’ | ||||||
| tetM | F:5’AGTGGAGCGATTACAGAA-3’ | 158bp | ||||
| R:5’CATATGTCCTGGCGTGTCTA-3’ | ||||||
| msrA | F:5’GAAGCACTTGAGCGTTCT-3’ | 287 bp | 94˚C for 4 min | 30 cycles of 94˚C-1 min, 50˚C-30 sec, 72 ˚C- 30 sec | 72 ˚C for 5 min | 13 |
| R:5’CCTTGTATCGTGTGATGT-3’ | ||||||
| dfrA | F:5’CTCACGATAAACAAAGAGTCA–3’ | 201 bp | ||||
| R:5’CAATCATTGCTTCGTATAACG – 3’ | ||||||
| mupA | F:5’TATATTATGCGATGGAAGGTTGG-3′ | 456bp | 94˚C for 2min | 30 cycles of 94˚C-45 sec, 53˚C-30 sec, 72 ˚C-45 sec | 72 ˚C for 2 min | 14 |
| R:5’AATAAAATCAGCTGGAAAGTGTTG-3′ | ||||||
| fusB | F:5’CCGTCAAAGTTATTCAATCG 3’ | 496bp | 94˚C for 2min | 30 cycles of 94˚C-45 sec, 53˚C-30 sec, 72 ˚C -45sec | 72 ˚C for 2 min |
15 |
| R:5’ACAATGAATGCTATCTCGACA 3’ | ||||||
| fusC | F:5’GGACTTTATTACATCGATTGAC 3’ | 128bp | ||||
| R:5’CTGTCATAACAAATGTAATCTCC 3’ | ||||||
| fusD | F:5’AATTCGGTCAACGATCCC 3’ | 525bp | ||||
| R:5’GCCATCATTGCCAGTACG 3’ | ||||||
Statistics
GraphPad Prism version 9 was employed to perform Fischer’s exact test. The association between antibiotic resistance and its respective resistance determinants was tested (p ≤ 0.05 was considered statistically significant).
All the phenotypically identified S. haemolyticus isolates (n=104) were confirmed by the presence of mvaA gene.
Phenotypic Screening of Antibiotic Resistance
(i) Antibiotic Susceptibility Testing
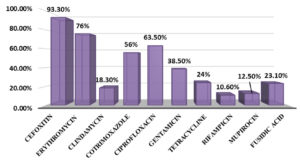
To vancomycin and linezolid, 100% susceptibility was shown by all tested isolates. The highest resistance (n=79, 76%) was observed for erythromycin, followed by ciprofloxacin (n=66, 63.5%) and cotrimoxazole (n=58, 55.8%). Relatively lower level of resistance was observed for gentamicin (n=40, 38.5%), followed by tetracycline (n=25, 24%), fusidic acid (n=24, 23.1%), clindamycin (n=19, 18.3%), mupirocin (n=12, 11.5%) and rifampicin (n=11, 10.6%). The overall antibiotic resistance profiles of the isolates are given in Figure 1.
(ii) Methicillin Resistance
The cefoxitin disc diffusion result revealed that majority of S. haemolyticus isolates were resistant to methicillin (n=97, 93.3%).
(iii) Detection of Inducible and Constitutive Clindamycin Resistance
Nineteen of the 104 isolates were non-susceptible to clindamycin, of which 14 were resistant, and the remaining five showed intermediate susceptibility. Inducible clindamycin resistance (iMLSB) was observed in eight isolates, and the remaining 11 isolates showed constitutive resistance (cMLSB).
Genotypic Screening of Antibiotic Resistant Genes
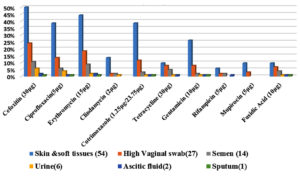
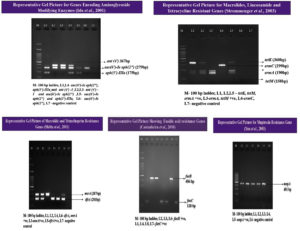
S. haemolyticus isolates (n=97, 93.3%) expressed the mecA gene, indicating resistance to methicillin. A high number of isolates were non-susceptible to erythromycin (n=79), of which 40 isolates (38.5%) were positive for the ermC gene and 31.7% (n= 33) were positive for the msrA gene. Five isolates (4.8%) contained a combination of the msrA and ermC genes, whereas one isolate (1%) showed a combination of the msrA with ermA genes. Non-susceptibility to cotrimoxazole was observed in 58 isolates (50: resistance, 8: intermediate resistance). The trimethoprim resistance-encoding gene dfrA was present in 54 isolates (52%). PCR detection of aminoglycoside modifying enzymes was performed for all gentamicin non-susceptible isolates (28: resistance, 12: intermediate resistance). A combination of the aac(6’)-Ie-aph(2’’) and aph(3’)-IIIa genes was detected in 28 isolates (26.9%), and the aac(6’)-Ie-aph(2’’) gene alone was observed in four isolates (3.8%). Nineteen isolates were resistant and six showed intermediate resistance to tetracycline (n=25, 24%), of which both genes were present in one of the isolates (1%). The tetK gene alone was present in 18 isolates (17.3%), and the tetM gene alone was present in one isolate (1%). Fusidic acid resistance was phenotypically observed in 24 isolates, of which 12 (11.5%) were positive for the fusB gene, and the remaining 12 (11.5%) were positive for the fusC gene; however, the fusD gene was absent. High level of mupirocin resistance was observed in 13 isolates; all 13 isolates (12.5%) were positive for the mupA gene. The representative gel pictures of the resistance genes screened in the present study are given in Figure 3. The complete antibiotic resistance profiles of the study isolates with both resistant phenotypes and genotypes are listed in Table 3. The correlation between the isolate source and antibiotic resistance was also determined (Figure 2). Strains isolated from the skin and soft tissue infections exhibited a comparatively high percentage of resistance to all antibiotics, followed by isolates from genital tract samples, such as high vaginal swab and semen, which showed increased antibiotic resistance. The sample-wise distribution of antibiotic resistance and its determinants is given in Figure 2 and Table 4 and 5.
Table (3):
Antibiotic resistance data comparing resistant phenotypes and genotypes (n-104).
| Antibiotic | Phenotypic Resistance | Genotypic Resistance {Respective Genes- No. (%)} | ||
|---|---|---|---|---|
| No. of Non-Susceptible Isolates (N-104) | (R- resistant, I- intermediate susceptibility) | (%) |
||
| Cefoxitin | 97 | R-97 | 93.3% | mecA– 97 (93.3%) |
| Erythromycin | 79 | R-70, I-9 | 76% | ermC– 40(38.5%) msrA- 33(31.7%) msrA+ermC- 5(4.8%) msrA+ermA- 1(1%) |
| Cotrimoxazole | 58 | R-50, I-8 | 56% | dfrA -54 (50.5%) |
| Gentamicin | 40 | R-28, I-12 | 38.5% | aac – 4(3.8%) aac+aph -28 (26.9%) |
| Tetracycline | 25 | R-19, I-6 | 24% | tetK -18 (17.3%) tetM– 1 (1%) tetK+tetM– 1(1%) |
| Mupirocin | 13 | R-13 | 12.5% | mupA -13 (12.5%) |
| Fusidic acid | 24 | R-24 | 23.1% | fusB – 12(11.5%) fusC – 12(11.5%) |
Table (4):
Sample wise antibiotic resistance profile.
N (%) |
Skin & soft tissues |
High Vaginal swab |
Semen |
Urine |
Ascitic fluid |
Sputum |
|---|---|---|---|---|---|---|
Cefoxitin |
52(50%) |
25(24%) |
11(10.6%) |
6(5.8%) |
2(1.9%) |
1(1%) |
Ciprofloxacin |
40(38.5%) |
14(13.5%) |
6(5.8%) |
4(3.8%) |
1(1%) |
1(1%) |
Erythromycin |
46(44.2%) |
19(18.3%) |
9(8.7%) |
2(1.9%) |
2(1.9%) |
1(1%) |
Clindamycin |
14(13.5%) |
2(1.9%) |
2(1.9%) |
1(1%) |
– |
– |
Cotrimoxazole |
40(38.5%) |
12(11.5%) |
3(2.9%) |
1(1%) |
1(1%) |
1(1%) |
Tetracycline |
10(9.6%) |
8(7.7%) |
5(4.8%) |
1(1%) |
1(1%) |
– |
Gentamicin |
27(26%) |
8(7.7%) |
2(1.9%) |
1(1%) |
1(1%) |
1(1%) |
Rifampicin |
6(5.8%) |
2(1.9%) |
2(1.9%) |
– |
1(1%) |
– |
Mupirocin |
10(9.6%) |
3(2.9%) |
– |
– |
– |
– |
Fusidic Acid |
10(9.6%) |
7(6.7%) |
4(3.8%) |
1(1%) |
1(1%) |
1(1%) |
Table (5):
Sample wise distribution of antibiotic resistance determinants.
| CX (n-97) | GEN (n-40) | COT (n-58) | ERY (n-79) | TET (n-25) | MUP (n-13) | FUS (n-24) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | mecA | aac | aac + aph | dfrA | msrA | ermC | msrA+ ermC | msrA+ ermA | tetK | tetM | tetK+ tetM | mupA | fusB | fusC |
| Source of Infection | ||||||||||||||
| Skin &soft tissues (54) | 52 (50%) | 4 (3.8%) | 15 (14.4%) | 29 (27.9%) | 25 (24%) | 28 (26.9%) | 3 (2.9%) | 1 (1%) | 6 (5.8%) | – | 1 (1%) | 10 (9.6%) | 4 (3.8%) | 6 (5.8%) |
| High Vaginal swab(27) | 25 (24%) | – | 8 (7.7%) | 12 (11.5%) | 4 (3.8%) | 6 (5.8%) | 1 (1%) | – | 7 (6.7%) | – | – | 3 (2.9%) | 3 (2.9%) | 4 (3.8%) |
| Semen (14) | 11 (10.6%) | – | 2 (1.9%) | 8 (7.7%) | 2 (1.9%) | 3 (2.9%) | – | – | 4 (3.8%) | 1 (1%) | – | – | 2 (1.9%) | 2 (1.9%) |
| Urine(6) | 6 (5.8%) | – | 1 (1%) | 2 (1.9%) | 1 (1%) | 1 (1%) | – | – | 1 (1%) | – | – | – | 1 (1%) | – |
| Ascitic fluid(2) | 2 (1.9%) | – | 1 (1%) | 2(1.9%) | 1 (1%) | 1 (1%) | 1 (1%) | – | – | – | – | – | 1 (1%) | – |
| Sputum(1) | 1 (1%) | – | 1 (1%) | 1(1%) | – | 1 (1%) | – | – | – | – | – | – | 1 (1%) | – |
| TOTAL (n-104) | 97 | 4 | 28 | 54 | 33 | 40 | 5 | 1 | 18 | 1 | 1 | 13 | 12 | 12 |
[CX-cefoxitin, GEN- gentamicin, COT-cotrimoxazole, ERY-erythromycin, TET-tetracycline, MUP-mupirocin, FUS-fusidic acid]
Statistical Analysis
No significant difference was observed between the antibiotic resistance and its determinants.
S. haemolyticus has been well known for its resistance to multiple antibiotics, which is also evident from the fact that it acquired methicillin resistance much earlier than other species of staphylococci.1 In this study, 93.3% of the isolates were methicillin resistant, and all the resistant isolates exhibited the mecA gene; however, in the study conducted by Barros et al.,4 among 64 methicillin-resistant S. haemolyticus isolates, 87% showed the mecA gene. In another study by Silva et al.,16 the mecA gene was present in 26 of 27 methicillin-resistant isolates. This proves that the phenotypic method cefoxitin disc diffusion is economical and can be reliably performed in a limited setup for surveillance of methicillin resistance.
Identification of a high percentage (85%) of multi-resistant strains was consistent with the results of other studies. Multidrug-resistant (MDR) strains in the study were defined as those “acquired non-susceptibility to at least one agent in three or more antimicrobial categories” (Magiorakos et al.).17 S. haemolyticus genome undergoes constant rearrangements, which is attributable to its multidrug resistance.
Indiscriminate and inappropriate use of broad-spectrum antibiotics has significantly increased the incidence of antibiotic resistance. This was reflected in the study results. Erythromycin non-susceptibility was observed in the maximum number of isolates (76%), followed by ciprofloxacin (63.5%) and cotrimoxazole (55.8%), the three being the most commonly prescribed broad-spectrum drugs in clinical setting. Surprisingly, last resort and the least prescribed drugs such as vancomycin and linezolid have shown 100% susceptibility. Similar results were reported by Krzyminska et al.18
MLS antibiotics, though chemically different, have similar resistance mechanism of ribosomal modification encoded by the erythromycin ribosome methylation (erm) gene.19,20 MLS antibiotics are clinically significant in the treatment of Gram-positive infections. Hence, cross-resistance between them is a clinical concern.21 In this study, among erythromycin-resistant isolates, the MSB phenotype was predominant (n=60, 57.7%), followed by cMLSB (n=11, 10.6%) and iMLSB (n=8, 7.7%). Furthermore, MLS phenotypes are considered to vary according to the geographic location. Hence, when analysing a similar Indian study by Manoharan et al.,5 on isolates from southern India, mainly Puducherry, cMLSB and MSB phenotypes had almost the same predominance (42.5% and 40.3%, respectively), whereas the MSB phenotype was predominant in the present study from Chennai.
Trimethoprim resistance is either chromosomally mediated that occurs due to mutations in the dfrG gene encoding dihydrofolate reductase (DHFR), the enzyme involved in the folate pathway, or plasmid mediated that occurs due to variants of DHFR having low affinity for trimethoprim.22 These DHFR variants are encoded by the dfrA, dfrD and dfrK genes, of which the dfrA gene is the most common. In the present study, 54/58 cotrimoxazole-resistant isolates exhibited the dfrA gene. The results were in concordance with the study by Aggarwal et al.,23; they screened three trimethoprim resistance genes from S. aureus isolates, of which the majority of isolates (45/74) carried the dfrA gene. In contrast, Manoharan et al.5 reported that among S. haemolyticus study isolates, 89.7% of cotrimoxazole-resistant isolates were dfrG-positive, and the dfrA gene in combination with other genes, including dfrD and dfrG, was present only in 2% of the isolates.
Aminoglycoside resistance in staphylococci is due to target site modification, leading to inactivation of the drug caused by aminoglycoside modifying enzymes.24 Plasmid mediated genes {aac(6’)-Ie-aph(2’’), aph(3’)-IIIa and ant(4′)} encoding three commonly found aminoglycoside modifying enzymes {AAC(6’)/APH(2’), APH(3’)-III, and ANT (4’)-I, respectively} were screened in this study. A high number of aminoglycoside-resistant isolates (26.9%) exhibited a combination of the aac(6’)-Ie-aph(2’’) and aph(3’)–IIIa genes in the present study. These findings disagree with those of other published studies that revealed the presence of aac(6’)-Ie-aph(2’’) alone rather than in combination. Both studies showed the lowest prevalence of the ant(4′) gene, whereas it was completely absent in the present study.25,18
Tetracycline resistance in staphylococci is either due to active efflux by acquiring the plasmid-mediated genes tetK and tetL or the chromosomal resistance genes tetM and tetO.26 In this study, the most frequently observed genes, tetK and tetM, were screened. Among the 25 tetracycline non-susceptible isolates, 72% were positive for the tetK gene. Unlike the findings of the present study, the tetM gene was predominant (67%) and the tetK gene was present in only 33% of the isolates in the study conducted by Duran et al.,27 However, the findings of the study by Manoharan et al.,5 were similar to those of the present study, with 91.5% prevalence of the tetK gene. Apart from the resistant isolates, susceptible isolates exhibiting resistance genes were also observed in both previous studies but was absent in the present study.
The most common resistance mechanism to fusidic acid is protecting the target site by the genes encoding the fusB family of proteins, thereby preventing the translocation of elongation factor G (EF-G) from the ribosome, leading to inhibition of protein synthesis. Casanteira et al.,18 compared the occurrence rates of fusidic acid resistance in Australia, Canada and the USA, and observed that the prevalence of fusidic acid resistance in CoNS was the highest in Canada (20%), followed by Australia (10.8%) and the USA (7.2%). In this study, the occurrence of fusidic acid resistance in Chennai, southern India, was 23% among S. haemolyticus study isolates. Half of the isolates exhibited prevalence of the fusB gene, and the remaining 50% exhibited the fusC gene. However, other studies have demonstrated a higher prevalence of the fusB gene than that of the fusC gene.15,28
Mupirocin is a bacteriostatic antibiotic that inhibits protein synthesis. Among the two phenotypes, high level of resistance is mediated by plasmid carrying the iles2 or mupA gene that encodes a novel tRNA synthetase. Among the study isolates, 12.5% exhibited high-level phenotypic resistance to mupirocin, all of which carried the mupA gene. These findings are consistent with those of other studies.29,30 Universal methicillin-resistant Staphylococcus aureus (MRSA) decolonisation protocol followed in hospitals is an important reason for increased resistance to mupirocin. Thus, stabilising the use of mupirocin with proper surveillance and target-based decolonisation may be of great help in controlling mupirocin resistance.
The isolate source was correlated with the resistance phenotypes and genotypes. It is well known that staphylococci normally inhabit the skin and mucous membranes in humans. Hence, the predominant S. haemolyticus isolates having the highest resistance to various antibiotics were from the skin and soft tissues. These findings were consistent with those of Palestine and Ethiopia.31,32 Interestingly, all resistant genotypes and their combinations were observed in isolates from the skin and soft tissues in the present study. In addition to skin and soft tissue infection samples, genital tract samples, such as high vaginal swab and semen, also exhibited high level of antibiotic resistance. Other samples (urine, ascitic, and sputum) were low in number to draw conclusions.
A high percentage of antibiotic resistance in opportunistic pathogens such as S. haemolyticus is a concern, as it may lead to treatment failure, prolonged hospital stay and increased mortality rate. In addition, there is a greater risk of disseminating resistance genes to other virulent species of staphylococci, making them increasingly arduous in hospital setup.
ACKNOWLEDGMENTS
The authors would like to thank University of Madras for rendering instrumentation facilities, Indian Council of Medical Research for the Financial support and ESIC Hospital, K.K. Nagar for providing clinical isolates for the study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
The study was financially supported by the Indian Council of Medical Research, with the Senior Research Fellowship (OMI-Fellowship/19/2018-ECD-I).
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Czekaj T, Ciszewski M, Szewczyk EM. Staphylococcus haemolyticus – an emerging threat in the twilight of the antibiotics age. Microbiology. 2015;161(11):2061-2068.
Crossref - Cavanagh JP, Hjerde E, Holden MTG, Kahlke T, Klingenberg C, Flægstad T, Parkhill J, Bentley SD, Sollid JUE. Whole-genome sequencing reveals clonal expansion of multiresistant Staphylococcus haemolyticus in European hospitals. Journal of Antimicrobial Chemotherapy. 2014;69(11):2920-2927.
Crossref - Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui L, Takahashi M, Ankai A, Baba S- ichi, Fukui S, Lee JC, Hiramatsu K. Whole-Genome Sequencing of Staphylococcus haemolyticus Uncovers the Extreme Plasticity of Its Genome and the Evolution of Human-Colonizing Staphylococcal Species. Journal of Bacteriology. 2005;187(21):7292-7308.
Crossref - Barros EM, Ceotto H, Bastos MCF, Dos Santos KRN, Giambiagi-deMarval M. Staphylococcus haemolyticus as an Important Hospital Pathogen and Carrier of Methicillin Resistance Genes. Journal of Clinical Microbiology. 2011;50(1):166-168.
Crossref - Manoharan M, Sistla S, Ray P. Prevalence and Molecular Determinants of Antimicrobial Resistance in Clinical Isolates of Staphylococcus haemolyticus from India. Microbial Drug Resistance. 2021;27(4):501-508.
Crossref - Koneman EW. Koneman. Lippincott Williams & Wilkins; 2006.
- Bauer AW, Kirby WM., Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. American journal of clinical pathology. 1966; 45(4): 493–496.
- Performance Standards for Antimicrobial Susceptibility Testing, Twenty-Fifth Informational Supplement. CLSI document M100-S25, Wayne, PA: Clinical and Laboratory Standards Institute. CLSI 2015.
- Pereira EM, Schuenck RP, Malvar KL, et al. Staphylococcus aureus, Staphylococcus epidermidis and Staphylococcus haemolyticus: Methicillin-resistant isolates are detected directly in blood cultures by multiplex PCR. Microbiological Research. 2010;165(3):243-249.
Crossref - Abimanyu N. Use of Triplex PCR for Rapid Detection of PVL and Differentiation of MRSA from Methicillin Resistant Coagulase Negative Staphylococci. Journal of clinical and diagnostic research. Published online 2013.
Crossref - Ida T, Okamoto R, Shimauchi C, Okubo T, Kuga A, Inoue M. Identification of Aminoglycoside-Modifying Enzymes by Susceptibility Testing: Epidemiology of Methicillin-Resistant Staphylococcus aureus in Japan. Journal of Clinical Microbiology. 2001;39(9):3115-3121.
Crossref - Strommenger B, Kettlitz C, Werner G, Witte W. Multiplex PCR Assay for Simultaneous Detection of Nine Clinically Relevant Antibiotic Resistance Genes in Staphylococcus aureus. Journal of Clinical Microbiology. 2003;41(9):4089-4094.
Crossref - Shittu AO, Okon K, Adesida S, et al. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiology. 2011;11(1):92.
Crossref - Yun H-J . Prevalence and mechanisms of low- and high-level mupirocin resistance in staphylococci isolated from a Korean hospital. Journal of Antimicrobial Chemotherapy. 2003;51(3):619-623.
Crossref - Castanheira M, Watters AA, Mendes RE, Farrell DJ, Jones RN. Occurrence and molecular characterization of fusidic acid resistance mechanisms among Staphylococcus spp. from European countries (2008). Journal of Antimicrobial Chemotherapy. 2010;65(7):1353-1358.
Crossref - Silva PV, Cruz RS, Keim LS, et al. The antimicrobial susceptibility, biofilm formation and genotypic profiles of Staphylococcus haemolyticus from bloodstream infections. Memórias do Instituto Oswaldo Cruz. 2013;108(6):812-813.
Crossref - Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268-281.
Crossref - Krzymińska S, Szczuka E, Dudzińska K, Kaznowski A. Virulence and the presence of aminoglycoside resistance genes of Staphylococcus haemolyticus strains isolated from clinical specimens. Antonie van Leeuwenhoek. 2015;107(4):857-868.
Crossref - Schmitz F-J, Verhoef J, Fluit AC, The Sentry Participants Group. Prevalence of resistance to MLS antibiotics in 20 European university hospitals participating in the European SENTRY surveillance programme. Journal of Antimicrobial Chemotherapy. 1999;43(6):783-792.
Crossref - Lim J-A. Prevalence of resistance to macrolide, lincosamide and streptogramin antibiotics in Gram-positive cocci isolated in a Korean hospital. Journal of Antimicrobial Chemotherapy. 2002;49(3):489-495.
Crossref - Szemraj M, Czekaj T, Kalisz J, Szewczyk EM. Differences in distribution of MLS antibiotics resistance genes in clinical isolates of staphylococci belonging to species: S. epidermidis, S. hominis, S. haemolyticus, S. simulans and S. warneri. BMC Microbiology. 2019;19(1).
Crossref - Vickers AA, Potter NJ, Fishwick CWG, Chopra I, O’Neill AJ. Analysis of mutational resistance to trimethoprim in Staphylococcus aureus by genetic and structural modelling techniques. Journal of Antimicrobial Chemotherapy. 2009;63(6):1112-1117.
Crossref - Aggarwal S, Jena S, Panda S, et al. Antibiotic Susceptibility, Virulence Pattern, and Typing of Staphylococcus aureus Strains Isolated From Variety of Infections in India. Frontiers in Microbiology. 2019;10.
Crossref - Doi Y, Wachino J, Arakawa Y. Aminoglycoside Resistance. Infectious Disease Clinics of North America. 2016;30(2):523-537.
Crossref - Perumal N, Murugesan S, Krishnan P. Distribution of genes encoding aminoglycoside-modifying enzymes among clinical isolates of methicillin-resistant staphylococci. Indian Journal of Medical Microbiology. 2016;34(3):350-352.
Crossref - Grossman TH. Tetracycline Antibiotics and Resistance. Cold Spring Harbor Perspectives in Medicine. 2016;6(4):a025387.
Crossref - Duran N, Ozer B, Duran GG, Onlen Y, Demir C. Antibiotic resistance genes & susceptibility patterns in staphylococci. The Indian journal of medical research. 2012; 135(3): 389–396.
- Hung W-C, Chen H-J, Lin Y-T, et al. Skin Commensal Staphylococci May Act as Reservoir for Fusidic Acid Resistance Genes. de Lencastre H, ed. PLOS ONE. 2015;10(11):e0143106.
Crossref - Bathoorn E, Hetem DJ, Alphenaar J, Kusters JG, Bonten MJM. Emergence of High-Level Mupirocin Resistance in Coagulase-Negative Staphylococci Associated with Increased Short-Term Mupirocin Use. Journal of Clinical Microbiology. 2012;50(9):2947-2950.
Crossref - Rudresh MS, Ravi GS, Motagi A, Alex AM, Sandhya P, Navaneeth BV. Prevalence of Mupirocin Resistance Among Staphylococci, its Clinical Significance and Relationship to Clinical Use. Journal of Laboratory Physicians. 2015;7(02):103-107.
Crossref - Al Laham DN, Abou Elkhair E, Bashir A, Abdelateef N. Resistance profiles and biofilm formation of coagulase negative staphylococci isolated from clinical specimens in a tertiary care hospital in Palestine. The International Arabic Journal of Antimicrobial Agents. 2017;7(3).
Crossref - Deyno S, Fekadu S, Seyfe S. Prevalence and antimicrobial resistance of coagulase negative staphylococci clinical isolates from Ethiopia: A meta-analysis. BMC Microbiology. 2018;18(1).
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.