ISSN: 0973-7510
E-ISSN: 2581-690X
This study investigated the bacteriological and the hygienic properties of some commonly sold canned milk and determined the effectiveness of pasteurization of milk contaminated with bacterial isolates spectrophotometrically. In this study, Escherichia coli, Streptococcus spp., Enterococcus aerogenes, Proteus spp., Clostridium spp., Lactobacillus spp. and Staphylococcus aureus were isolated from the milk samples. The bacterial load of the milk samples ranged from 5 x 105 to 22 x 105 cfu/ml. The methylene reduction time indicating the hygienic quality of each milk sample varied from one milk to the other. While NNM and VSM samples not decolourized after 8 h were considered excellent and PNM and OFM decolourized methylene blue after 6 and 7 h respectively were considered good milk samples, CFM and TCM that decolourized methylene blue after 4 and 5 h respectively were considered fair milk samples. None of them was considered poor as they were not decolourized in less than 2 h. Pasteurizing milk contaminated with indigenous microflora (IMF), E. coli and Strep. pyogenes at 40oC, 50oC, 60oC and 70oC over a period of 30 min showed a varied degree of reduction in the absorbances and its effectiveness. A 10oC increase in temperature resulted in significant reduction in the absorbances obtained at different time intervals for all the milk samples. This investigation shows that pasteurizing milk with low heating would reduce the microbial loads of the milk samples. The method of pasteurizing milk in this study would be applicable in the household dairies where sophisticated equipment are not required to control microbial loads in milk.
Contamination, methylene blue reduction, indigenous microflora, pasteurization
Milk is a highly nourishing and important food with potential therapeutic properties. It is an all-inclusive food consumed in a variety of different products by millions of people. Rolf1 described milk as colloids of butterfat globules within a water-based fluid containing dissolved carbohydrates. While containing proteins, fat, sugar, vitamins and minerals2, 97–98% lipids in milk are triacylglycerols, small amounts of di- and monoacylglycerols, free cholesterol and cholesterol esters, free fatty acids and phospholipids3. The fat portion of the milk contains fat-soluble vitamins, linoleic and linolenic acid 4. Being a delicate agricultural product and an enrichment medium, its nutrient compositions easily support bacterial growth5-8. It may be contaminated from animal skin, environment, utensils and water used for adulteration9 and the quality of the contaminated milk may deteriorate.during transportation at ambient temperature10. Consequently, many bacterial strains including Listeria monocytogenes, Salmonella sp., Campylobacter sp., Staphylococcus aureus, Bacillus cereus, Escherichia coli and some species of Streptococcus and Micrococcus have been reported as pathogenic and spoilage bacteria in milk 11-16. While many contaminating bacteria spoil the milk and others cause the souring of milk and its products naturally, many are know to cause gastroenteritis and diarrhea17.
Although De Man et al.,18 and Kleter,19 had earlier reported that freshly harvested milk usually have a low microbial load with total count usually less than 103 cfu/ml, Chye et al.20 showed that the microbial load could increase when stored at ambient temperature for a long period of time. While the microbial content of milk determines its quality21,22, the presence of high microbial load in milk poses major economic loss for farmers and household dairies as milk price is determined by the value of the pathogenic bacteria count23,24. Since Vasavada25 considered milk as a major vehicle for transmitting infectious agents, Coorevits et al26 and FAO9 reported that bacterial contamination of milk could be from different sources. Consequentially, since careful hygiene is needed during processing to avoid microbial contamination and there is a significant threat to milk recipients who are vulnerable to food-borne infectious diseases due to low age and co-morbidities27, determining the bacteria load, characterizing the indigenous flora, pasteurizing and determining the absorbance rate of bacterial contaminated milk samples at different temperatures for varying durations and the potential of successfully practicing low scale pasteurization procedure becomes inevitable to ensure the quality and safety of milk consumed by the populace. This study was, therefore, aimed at investigating the bacterial contaminants and the effectiveness of pasteurizing milk contaminated with bacterial pathogens in a miniature experiment to show the possibility of pasteurizing milk by small scale farmers in a cost effective manner.
Source of samples
Five different evaporated canned milk and a soy milk sample were purchased randomly at different markets located in Ogun State, Nigeria. Each sample was labeled and surface sterilized with 70% ethanol before being opened with a heat sterilized tin cutter. Each milk sample was emptied in labeled sterilized autoclave-able plastic bottles. The milk samples were kept in ice packs before being kept in refrigerator at 3oC.
Total viable aerobic bacterial count
Total viable aerobic bacterial and coliform counts of the milk samples was determined as described by Collins et al28. Each of the samples was diluted tenfold in properly labeled 20 ml glass tubes containing 9 ml sterile normal saline as diluent. Each sample was aseptically diluted serially with separate sterile glass pipette (10 ml). One ml sample from tube dilution number 10-8, 10-7, 10-6, 10-5 and 10-4 was dispensed on surfaces of nutrient agar and Eosin Methylene Blue (EMB) agar plates in triplicate before being spread evenly with sterile glass spreader. After incubating each plate at 37oC for 48 h, total colony forming units (cfu) of aerobic bacterial and coliform counts were recorded from the plate having 30-300 colonies.
Isolation and identificantion of isolates from milk samples
Serially diluted milk samples were plated in duplicate in nutrient broth and McConkey broth using pour plate technique. Plates were incubated at 37oC for 24 – 48 h. Total viable counts were carried out on nutrient agar while the presence or absence of some organisms was done by plating on selective media. Pure cultures of discreet colonies were made by repeated sub-culturing on sterile nutrient agar with a sterile wire loop. Using overnight cultures, the pure strains were identified and confirmed using morphological, microscopy and biochemical tests following standard procedures described by Cowan et al.,29 and Cheesborough30. Results were analyzed using Bergy’s manual and other methods for bacterial identification31-33.
Methylene blue reduction test
Methylene blue reduction test (MBR) was conducted to determine the hygienic status of each of the milk samples34. One ml of methylene blue solution (1:25000) was transferred to labeled and sterile 20 ml screw capped test tube containing 10 ml of each of the milk samples. The tubes were capped and gently inverted three times to mix up the dye with each milk sample. Each of the tubes was incubated at 37oC and examined for discolourization at intervals up to 8 h. The time taken by the methylene blue in each milk sample to become colorless was recorded. The methylene blue reduction test depends upon the ability of bacteria in milk to grow and to consume the dissolved oxygen which reduces the oxidation-reduction potentials in the medium.
Pasteurization and Spectrophotometry at different temperature and varied time intervals
Pure strains of Escherichia coli and Streptococcus pyogenes were obtained from the Medical Laboratory Department of Babcock University Teaching Hospital, Ilishan Remo, Ogun State, Nigeria. The identity of these isolates were further confirmed by microbiological and biochemical tests. Overnight broth cultures of these bacteria were centrifuged at 1500 rpm for 5 min. The supernatants were discarded while the pellets were resuspended in normal saline. These were then centrifuged at 15000 rpm for 5 min and the supernatants were discarded again. The pellets were then resuspended in the normal saline. The absorbances of the bacterial suspensions were measured spectrophotometrically at a wavelength of 600 nm35. Dilutions of 106 cfu/ml of each each isolate from milk samples were prepared in triplicates. A predetermined volume of the diluted milk samples (9 ml) were pipette into the test tubes and 1 ml of adjusted E. coli and S. pyogenes suspensions was pipetted into two different sets of test tubes while the third set containing the indigenous flora (IMF) of the milk sample was not inoculated. The test tubes were then placed in the water bath at varying temperatures (40oC, 50oC, 60oC and 70oC). At specific time intervals of 10, 20 and 30 min, the absorbances of each contaminated milk sample were measured at a constant wavelength of 600 nm. The contaminated milk samples were later subcultured to check for the effectiveness of pasteurization procedure.
In this study, milk samples were tested for the presence of bacterial contaminants before being pasteurized. The pasteurized milk samples were then tested for methylene blue reduction to determine their hygienic status before being contaminated with the indigenous microbial flora (IMF), Escherichia coli and Streptococcus pyogenes. The bacteria isolated from the milk samples included Escherichia coli, Streptococcus spp., Enterococcus aerogenes, Proteus spp., Clostridium spp., Lactobacillus spp. and Staphylococcus aureus. The result showed that the isolates from the milk samples included Gram negative bacilli (GNB) 45.45%, Gram positive cocci (GPC) 36.36% and Gram positive bacilli (GPC) 18.18% (Table 1). The bacterial loads of the different milk samples were negligible and ranged from 5 x 105 to 22 x 105 cfu/ml (Table 2). The methylene reduction time indicating the hygienic quality of each milk sample varied from one milk to the other. While milk samples NNM and VSM were not decolourized after 8 h, others were decolourized between 4 and 7 h. The milk samples not decolourized were considered excellent. PNM and OFM decolourized after 6 and 7 h respectively were considered good milk samples while CFM and TCM decolourized after 4 and 5 h respectively were considered fair milk samples. None of the milk samples was considered poor as they were not decolourized in less than 2 h (Table 3).
Table (1):
Bacteria isolated from the different milk samples
Sample code |
Gram reactions |
Isolated bacteria |
|---|---|---|
NNM |
GNB |
Escherichia coli |
GPC |
Streptococcus spp. |
|
GNB |
Enterobacter aerogenes |
|
VSM |
GNB |
Proteus spp |
GPB |
Clostridium spp |
|
TCM |
GPB |
Lactobacillus spp |
CFM |
GPB |
Clostridium spp |
GPC |
Staphylococcus aureus |
|
PNM |
GNB |
Escherichia coli |
GPB |
Lactobacillus spp |
|
OFM |
GNB |
Proteus spp |
KEY: + Positive, – Negative, GPC – Gram Positive Cocci, GPB – Gram Positive Bacilli, GNB -Gram Negative Bacilli.
Table (2):
Results for Bacterial Count of the different milk samples
Sample Codes |
Number Of Colonies (cfu/ml) |
|---|---|
TCM |
6 x 105 |
NNM |
9 x 105 |
CFM |
19 x 105 |
OFM |
16 x 105 |
PNM |
22 x 105 |
VSM |
5 x 105 |
Table (3):
Results for methylene blue reduction test to determine the hygienic status of different milk samples
Sample Code |
Methylene Blue Reduction Time (h) |
Remark |
Class |
|---|---|---|---|
CFM |
4 |
fair |
3 |
TCM |
5 |
fair |
3 |
PNM |
6 |
good |
2 |
NNM |
Not Decolourized |
excellent |
1 |
OFM |
7 |
good |
2 |
VSM |
Not Decolourized |
excellent |
1 |
Key: Class 1 = > 8 h; Class 2 = < 8 h; Class 3 = < 6 h but not < 2 h.
After determining the initial microflora and the bacterial load of each milk sample, they were pasteurized before being contaminated with 106 cfu/ml of the indigenous microbial floral (IMF), E. coli and Strep. pyogenes. The results showed that a varied degrees of reduction in the absorbances were recorded for the contaminated milk samples pasteurized at 40oC, 50oC, 60oC and 70oC over a period of 30 min. With an increase in temperature of 10oC, there were significant reductions in the absorbances obtained at different time intervals for all the milk samples. The reductions in the absorbances of E. coli contaminated milk samples were higher than those obtained from IMF and S. pyogenes contaminated milk samples. Those of E. coli were followed by IMF contaminated milk samples while those of S. pyogenes contaminated milk samples were the least at the different temperature used in pasteurization. In all these contaminated milk samples, the absorbances were reduced as time and temperature were increased. At 0 min when pasteurized at 40oC, TCM containing IMF, E. coli and S. pyogenes had an absorbances of 0.552 nm, 0.578 nm and 0.544 nm respectively. After 30 min of pasteurization, the absorbances were reduced to 0.532 nm, 0.554 nm and 0.472 nm respectively. At 0 min when pasteurized at 50oC, the absorbances were reduced to 0.491 nm, 0.490 nm and 0.426 nm from 0.534 nm, 0.559 nm and 0.472 nm in TCM contaminated with IMF, E. coli and S. pyogenes respectively. After 30 min of pasteurizing at 60oC, the absorbance was reduced from 0.491 nm, 0.490 nm, and 0.426 nm at 0 min to 0.403 nm, 0.408 nm and 0.322 nm. After 30 min of pasteurizing at 70oC, the absorbances were reduced from 0.403 nm, 0.408 nm and 0.322 nm at 0 min to 0.262 nm, 0.283 nm and 0.221 nm. The same trend of reductions in the absorbances were obtained in the different milk samples contaminated with IMF, E. coli and S. pyogenes (Fig. 1 – 12). To ensure sterility, most of the contaminated milk subcultured on sterile nutrient showed no bacterial growth.
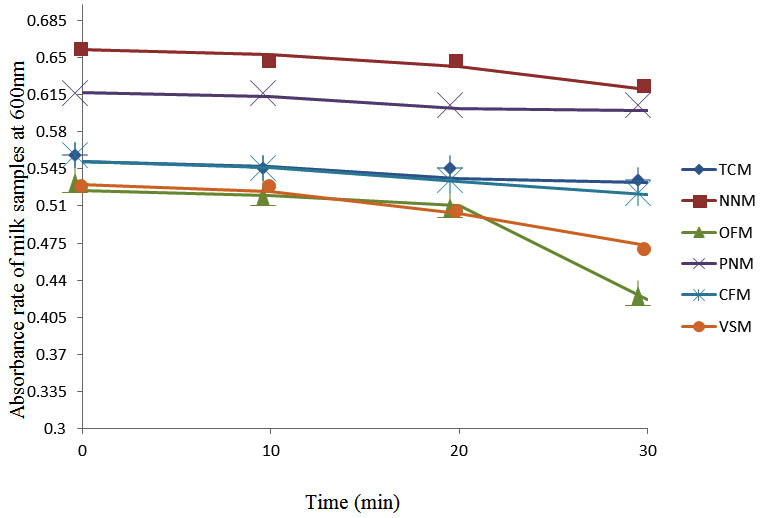
Fig. 1. Effect of pasteurization on milk samples contaminated with Indigenous Milk Flora (IMF) at 40°C at 600 nm
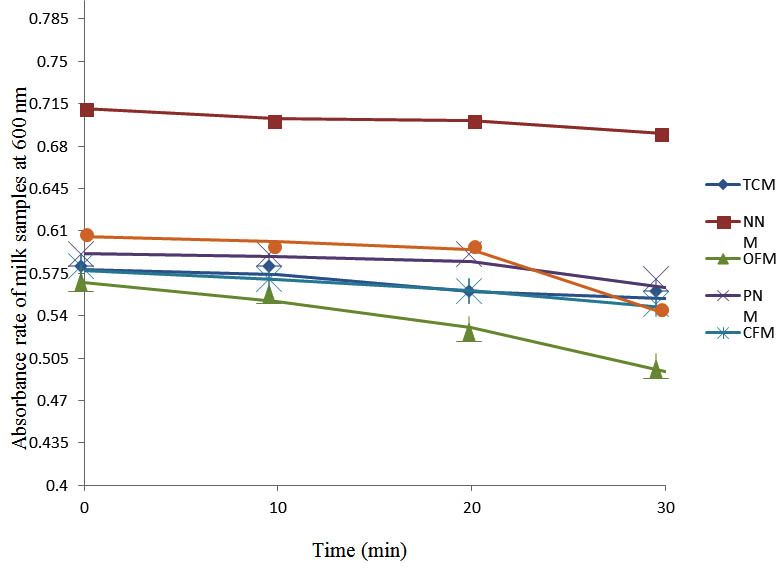
Fig. 2. Effect of pasteurization on milk samples contaminated with E. coli at 40°C at 600 nm
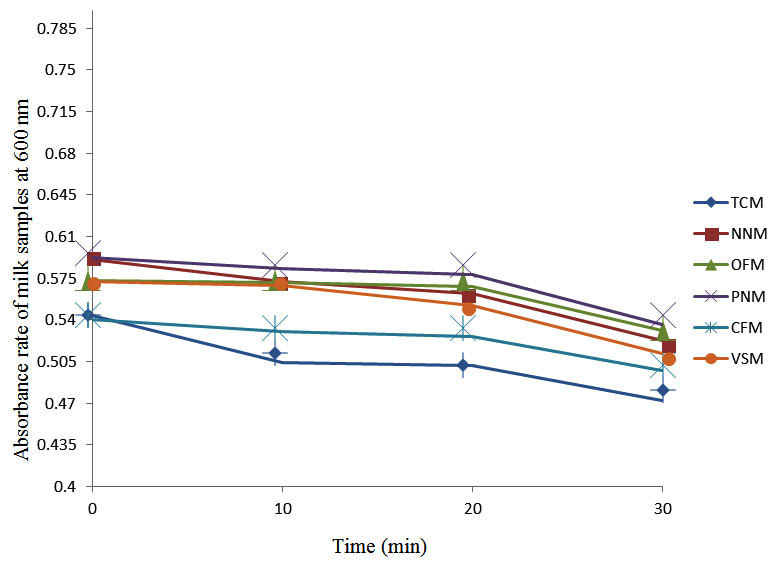
Fig. 3. Effect of pasteurization on milk samples contaminated with S. pyogenes at 40°C at 600 nm
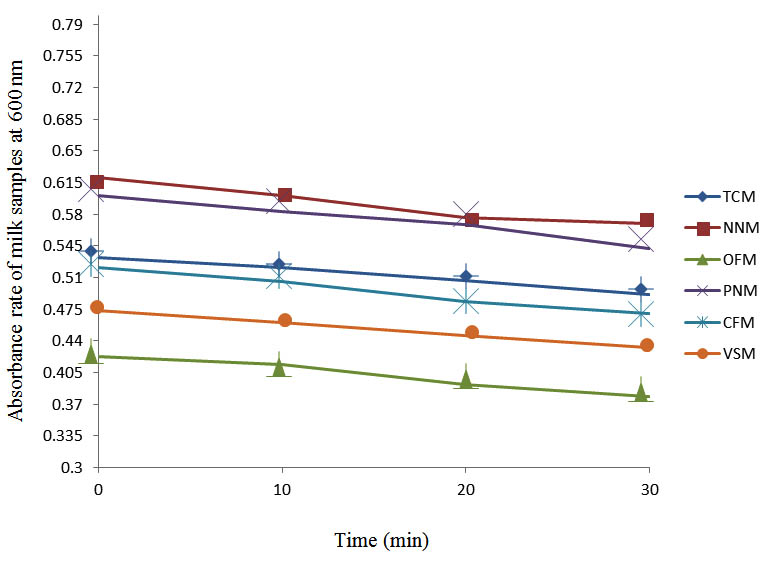
Fig. 4. Effect of pasteurization on milk samples contaminated with Indigenous Milk Flora (IMF) at 50°C at 600 nm
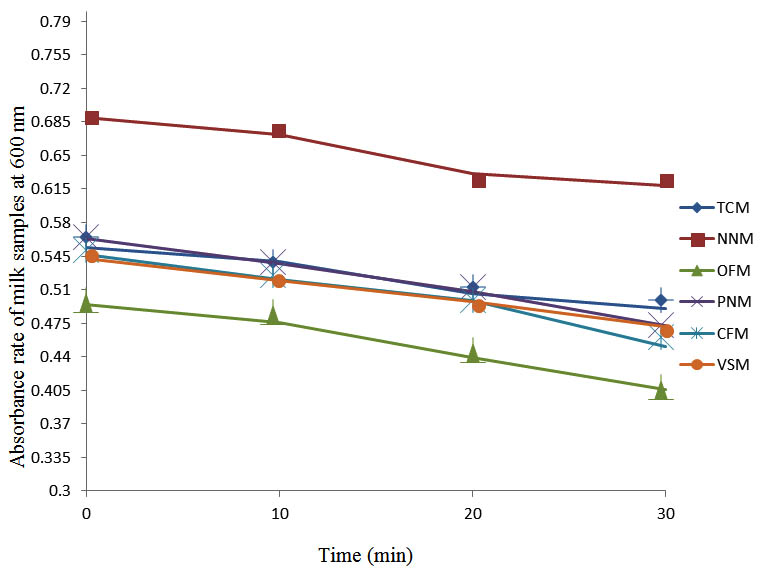
Fig. 5. Effect of pasteurization on milk samples contaminated with E. coli at 50°C at 600 nm
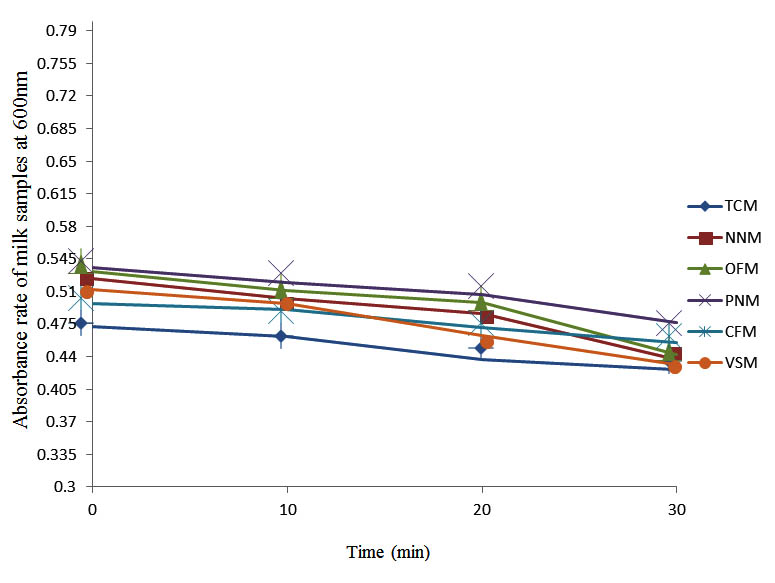
Fig. 6. Effect of pasteurization on milk samples contaminated with S. pyogenes at 50°C at 600 nm
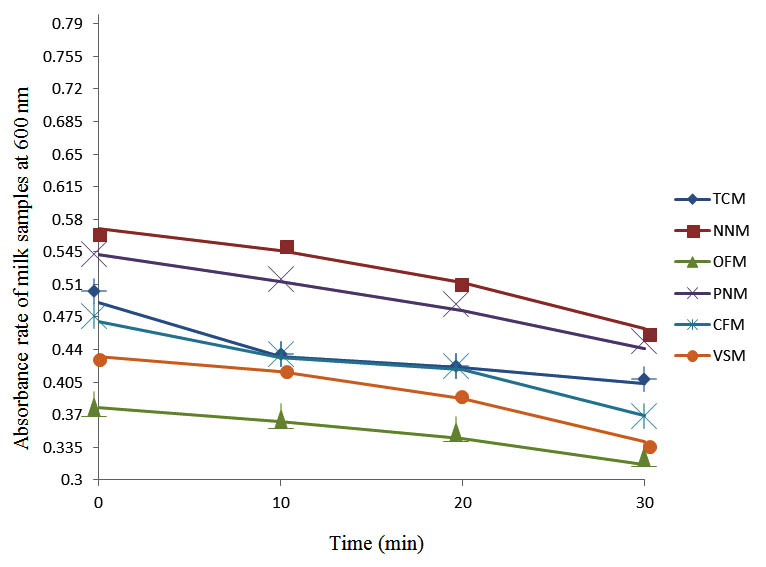
Fig. 7. Effect of pasteurization on milk samples contaminated with Indigenous Milk Flora (IMF) at 60°C at 600 nm
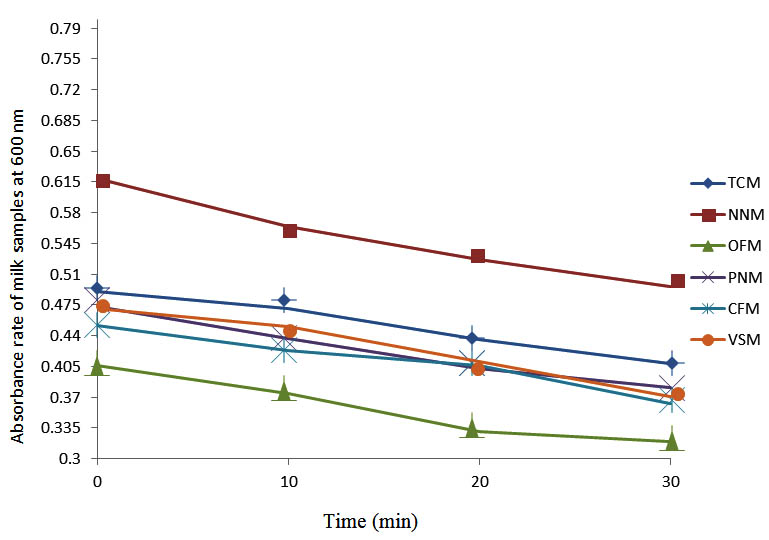
Fig. 8. Effect of pasteurization on milk samples contaminated with E. coli at 60°C at 600 nm
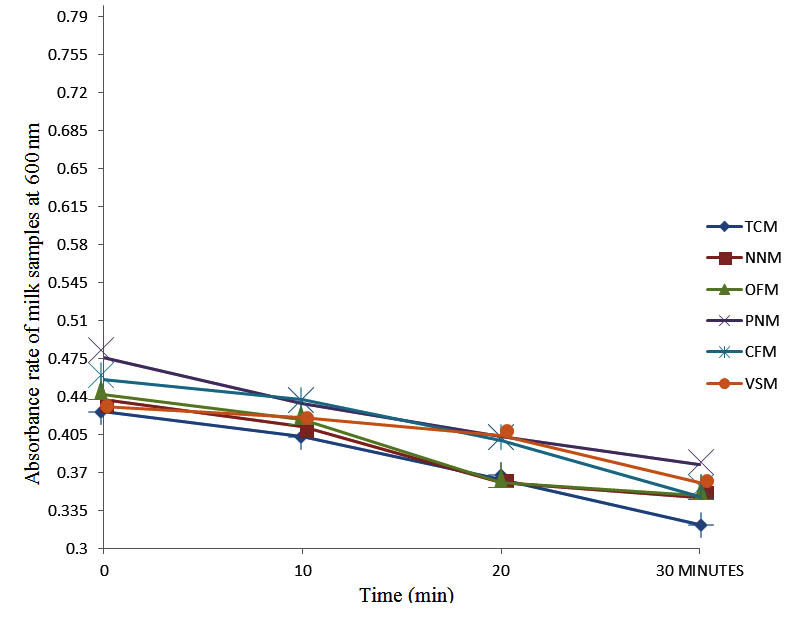
Fig. 9. Effect of pasteurization on milk samples contaminated with S. pyogenes at 60°C at 600 nm
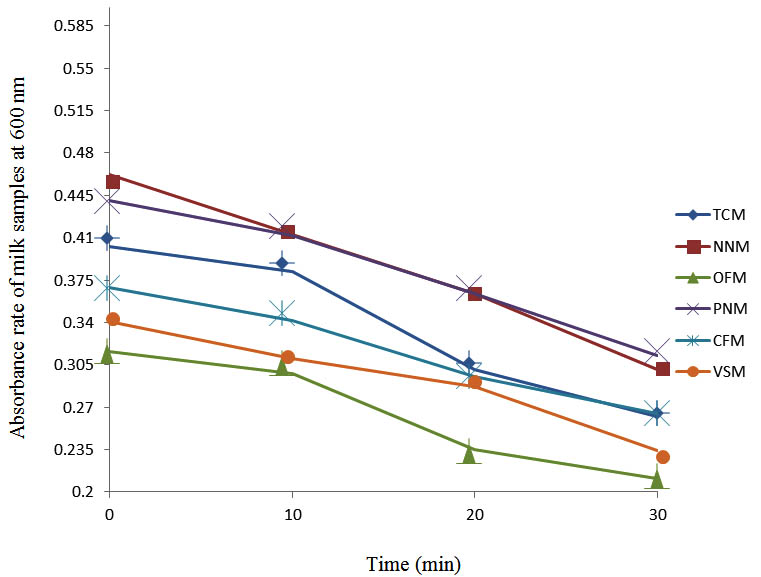
Fig. 10. Effect of pasteurization on milk samples contaminated with Indigenous Milk Flora (IMF) at70°C at 600 nm
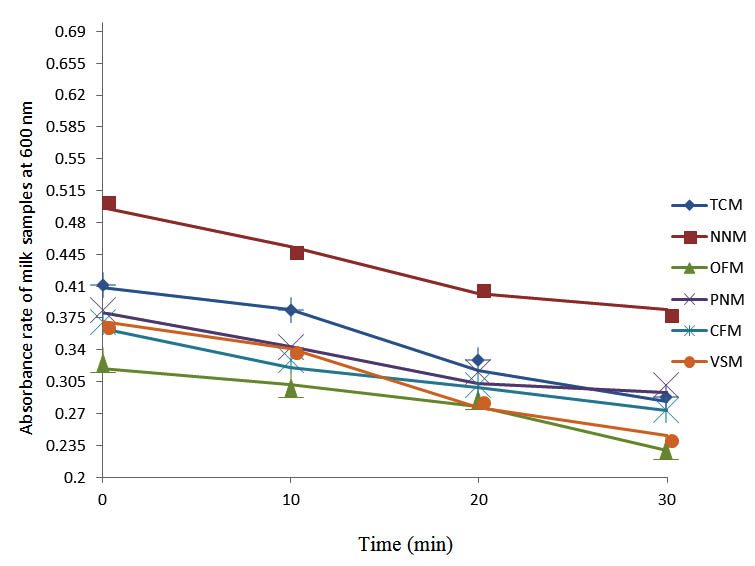
Fig. 11. Effect of pasteurization on milk samples contaminated with E. coli at 70°C at 600 nm
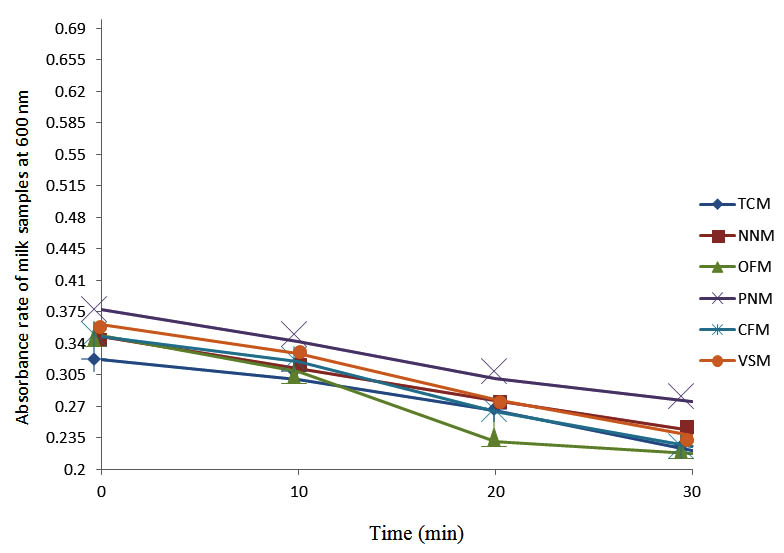
Fig. 12. Effect of pasteurization on milk samples contaminated with S. pyogenes at 70°C at 600 nm
Pasteurization is a moderate heat treatment of milk. It destroys disease producing organisms and limits fermentation in milk by heating milk to 63oC for 115 min while slow growing organisms and spore formers are hardly affected36. While pasteurization destroys many microorganisms in milk, improper handling after pasteurization could lead to its recontamination. Thus, the isolation of different strains of bacteria from the milk samples used in this study could have being through recontamination from pasteurization equipment prior packaging. Although pasteurization is not sterilization and meant to reduce the pathogens, the negligible level of microbial loads ranging from 5 x 105 to 22 x 105 cfu/ml indicated the effectiveness of the pasteurization methods employed by the various manufacturing companies. However, since the microbial content of milk is a major feature in determining its quality37,38, the methylene reduction time indicating the hygienic quality of each milk sample showed that NNM and VSM samples were excellent milk samples, PNM and OFM were considered good milk samples while CFM and TCM were considered fair milk samples.
However, since El-Mohandes et al.39 reported that 30% of contaminated milk samples in their study had more than 30,000 cfu/ml in which the highest infection was related to S. epidermidis, the presence of microbial contaminant in the samples showed that they are potential source of microbial infections depending on the virulence, pathogenicity and inoculums sizes. Though the microbial loads were negligible, that milk is nutritious and could support microbial growth indicated that the bacterial load of the indigenous microbial flora could increase under favourable conditions. Since poor milk hygiene have implications on the structure of milk, processing value, shelf life and edible food loss40 and indirectly on human health and bacterial contamination41-45, a high biological count in raw milk potent possible problems with product safety46.
In agreement with previous studies El-Mohandes et al37, Lindemann et al47 and Rozolen et al48, the isolation of Escherichia coli, Streptococcus spp., Enterococcus aerogenes, Proteus spp., Clostridium spp.,Lactobacillus spp. and Staphylococcus aureus from milk samples used have been reported. While El-Mohandes et al37, Lindemann et al.,47, Rozolen et al.,48 and Carroll et al.49 showed that frequently isolated microorganisms from milk samples included S. epidermidis, S. aureus, Enterobacter, Group B Streptococcus, Escherichia coli and Klebsiella, Karimi et al.50 reported Klebsiella, S. epidermidis, Enterobacter, Escherichia coli, Pseudomonas aeruginosa, Acinetobacter, S. saprophyticus, Citrobacter and S. aureus. To forestall outbreaks of infectious diseases from milk contaminated with these pathogens, several milk samples have be been pasteurized at varied temperature to ascertain its effectiveness in decontaminating milk samples. Wallner et al.51 showed that pasteurization is effective in destroying the bacteria in milk that cause tuberculosis, salmonellosis, diphtheria, typhoid fever and other illnesses without adversely affecting the milk’s nutritional content, flavor or quality. Grant et al.52 reported that milk contaminated with Mycobacterium paratuberculosis was decontaminated with a high-temperature and short-time pasteurization. While Orloff et al.53 reported that human immunodeficiency virus Type I was not recovered from human milk after pasteurization, Egbere et al.54 pasteurized kunun-zaki at 70oC for 30 min and discovered that the bacterial load was drastically reduced from 9.4 x 105 cfu/ml at 0 minutes to 3.1 x 101 cfu/ml after 30 min. At 15 min, most of the Streptococcus spp present in the sample had been killed.
From this study, the decreased absorbances of each milk sample contaminated with the indigenous milk flora (IMF), E. coli and S. pyogenes showed that the pasteurized milk contained very few organisms and agreed with the reports indicating that heating at 63°C for 30 min or 72°C for 15 min followed by quick cooling at 13°C destroys most milk-borne pathogens or reduces them to levels acceptable by international standards55-57. The spectrophotometric analysis of the pasteurized contaminated milk samples showed that all the samples obeyed Huygens’ principle stating that light passing through a suspension of microorganisms is scattered and the amount of scattered light is an indication of the biomass present in the samples57. The absorbance rates of all tested pasteurized milk samples was reduced because the concentration and number of organisms present in all the milk liquids were reduced by the pasteurization.
In conclusion, milk is one of the most consumed foods in the world and contained virtually all nutrients needed by man. If properly pasteurized, it should not contain contaminants, pathogenic organisms or coliforms indicating feacal contamination. This study indicates that, although not all milk samples were made sterile by the pasteurization, pasteurization of milk would reduce the number and concentration of organisms present in milk to levels acceptable by international community as most organisms present are killed by heating at 70°C for 30 min. The method of pasteurizing milk in water bath at varied temperature as used in this study would be applicable in the homestead where sufisticated equipment are not required to control microbial loads in milk.
- Rolf, J. Milk and Dairy Products. Ullmann’s Encyclopedia of Industrial Chemistry, Wiley-VCH, Weinheim. 2002; 16: 3-6.
- Komorowski, E.S., Early, R. Liquid milk and cream. In: The Technology of Dairy Products, Early, R. (Ed.) Blackie, London, 1992, pp. 305.
- Chen, H., O’Reilly, E., McCullough, M.L., Rodriguez, C., Schwarzschild, M.A., Calle, E.E., Thun, M.J., Ascherio, A. Consumption of Dairy Products and Risk of Parkinson’s Disease. Am. J. Epidemiol., 2006, 165(9): 998–1006.
- McGee, H., On Food and Cooking. 1st Edn., Scribner Press, ISBN 0-684-80001-2, New York, 2004,pp. 494.
- Byan, F.L. Epidmiology of milk-borne diseases. J. Food Prot., 1983; 46: 637-649.
- Bramley, A.J., McKinnon, C.H. The Microbiology of raw milk. In: Dairy Microbiology, I. (Ed.: Robinson, R.K.). London, New York, Elsevier Applied Science, 1990; pp: 171.
- Heeschen, Introduction In: Monograph on the significance of the pathogenic microorganisms in raw milk. International Dairy Federation Brussels, 1994, pp: 8-11.
- Ruegg, P.L. Practical food safety interventions for dairy production. J. Dairy Sci., 2003; 86: E1-E9.
- FAO. Milk Hygiene, In Milking, Milk Production Hygiene and Udder Health, FAO Animal Production and Health Papers-78, FAO Corporate Document Repository, 2008; 1-7 pp.
- Schmidt, G.H., Van Vleck, L.D. Principles of Dairy Science. Surjeet publications. New Delhi. 1982; 3-5 pp.
- Adesiyun, A.A., Stoute, S., David, B. Pre-processed bovine milk quality in Trinidad: Prevalence and characteristics of bacterial pathogens and occurrence of antimicrobial residues in milk from collection centers. Food Contr., 2007; 18: 312-320.
- Kagkli, D.M., Vancanneyt, M., Hill, C., Vandamme P., Cogan, T.M. Enterococcus and Lactobacillus contamination of raw milk in farm dairy environment. Int. J. Food Microbiol., 2007, 114: 243-251.
- Dudrikova, E., Polakova, L., Pukacova, J. Health and hygienic conditions of ewe‘s milk processing from the aspect of food safety. Potravinarstvo 2010; 4(3): 14-18.
- Fabianova, J., Duckova, V., Canigova, M., Krocko, M. Presence of Enterococci in cow milk and their antibiotic resistance. Potravinarstvo, 2011; 4(2), 17-21.
- Polakova, L., Dudrikova, E., Gallo, J. Presence of S. aureus and Enterococcus spp. in goat‘s cheese and their antibiotics resistance. Potravinarstvo, 2011; 5(3): 33-37.
- Zigo, F., Vasil, M., Kadasi, M., Elecko, J., Farkasova, Z. Bacteria Staphylococcus spp. isolated from mastitis of sheep their enterotoxigenic properties. Potravinarstvo, 2011; 5(4): 70-72.
- Oliver, S.P., Boor, K.J., Murphy, S.C., Murinda. S.E. Food safety hazards associated with consumption of raw milk. Foodborne Path. Dis., 2009; 6: 793-806.
- De Man, J.C., Rogosa, M., Sharpe, M.E. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol., 1960; 23: 130-135.
- Kleter, G. The bacterial flora in aseptically draw milk neth. Milk dairy J., 1975; 28: 220-237.
- Chye, F.Y., Abdullahb, A., Ayob, M.K. Bacteriological quality and safety of raw milk in Malaysia. Food Microbiol., 2004; 21: 535-541.
- Rogelj Mleko, I. In: Mikrobiologija zivil zivalskega izvora (Eds.: Bem, Z., Adamic, J., Zlender, B., Smole Mozina, S., Gasperlin, L.). Ljubljana, Biotehniska fakulteta, Oddelek za zivilstvo, 2003; pp: 515-538.
- Singh, V., Kaushal, S., Tyagi, A. and Sharma, P. Screening of bacteria responsible for the spoilage of milk. Journal of Chemical and Pharmaceutical Research 2011; 3: 348-350.
- Bonfoh, B., Wasem, A., Traore, A.N., Fane, A., Spillmann, H., Simb, C.F., Alfaroukh, I.O., Nicolet, J., Farahb, Z., Zinsstag, J. Microbiological quality of cow’s milk taken at different intervals from the udder to the selling point in Bamako (Mali). Food Contr., 2003; 14: 495-500.
- Verdier-Metz, I., Michel, V., Delbes, C., Montel, M.C. Do milking practices influence the bacterial diversity of raw milk. Food Microbiol., 2009; 26: 305-310.
- Vasavada, P.C. Pathogenic bacteria in milk: A review. J. Dairy Sci. 1988; 71: 2809-2816.
- Coorevits, A., De Jonghe, V., Vandroemme, J., Reekmans, R., Heyrman, J., Messens, W., De Vos, P., Heyndrickx, M. Comparative analysis of the diversity of aerobic-spore forming bacteria in raw milk from organic and conventional dairy farms. System. Appl. Microbiol., 2008; 31(2): 126-40.
- ANVISA, Curso Básico de Controle de Infecção Hospitalar. Caderno e Programa do Controle de Infecção Hospitalar. Brasília: Brazilian Ministry of Health. 2000; http://www.scielo.br/scielo.php?script=sci_nlinks&ref=000079&pid=S1413-8670200800030001100002&lng=en
- Collins, C.H., Lyne, P.M., Grange, J.M. Counting Microorganisms. In Microbiological Methods, 6th Edition, Butterworth/Heinemann 1989; pp. 127-140.
- Cowan, S.J., Steel, K.J. Cowan and Steel manual for identification of medical bacteria, 2nd edn Cambridge University Press, London, 1974; pp. 176-232.
- Cheesborough, M. Medical Laboratory Manual for Tropical Countries, II Micrbiology (ELBS), Butterworth, Kent, U.K, 2006; pp. 23-78.
- Barrow, G.I., Feltham, R.K. Cowan and Steel Manual for the identification of Medica Bacteria, 3rd Edn, Cambridge 1993.
- Da-Silva, Z.N., Da-Cunha, A.S., Lins, M.C., Carneiro, L.D.A M., Almeida, A.C.D.F., Queiroz, M.L.P. Isolation and serological identification of enteropathogenic Escherichia coli in pasteurized milk in Brazil. Revista de Saude Publica. 2001; 35(4): 375-379.
- Ellis, D.L., Goodacre, R. Detection, identification,a nd enumeration methods for spoilage yeasts. In; Food Spoilage organisms, Blackburn C de W (Ed)., CRC Press LLC. 2006; Pp:28-54.
- Chatterjee, S.N., Bhattacharjee, I., Chatterjee, S.K., Chandra, G. Microbiological examination of milk in Tarakeswar, India with special reference to coliforms. Afr. J. Biotechnol., 2006; 5(15): 1363–1385.
- Sutton, S. Measurement of cell concentration in suspension by optical density. J. Appl. Microbiol. Biotechnol., 2006; B: 252-259.
- Namminga, K. “Health Risks of Drinking Raw (Unpasteurized) Milk,” http://www.abs.sdstate.edu/flcs/ecoli/milk.htm, South Dakota State University, Brookings, SD (reviewed by E. Kim Cassel, SDSU Extension Dairy Specialist) 1999.
- Chandan, R.C., Kilara, A., Shan, N.P. Dairy Processing and Quality Assurance. Wiley-Blackwell, Ames, Iowa, USA., ISBN: 0-8138-2756-6, 2008; pp. 329.
- Tamime, A.Y. Milk Processing and Quality management. John Wiley & Sons in Blackwell Publishing Ltd., Chichester, UK., ISBN: 978-1-4051-4530-5, 2009; pp. 316.
- El-Mohandes, A.E., Schatz, V., Keiser, J.F., Jackson, B.J. Bacterial contaminants of collected and frozen human milk used in an intensive care nursery. Am. J. Infect. Contr., 1993; 21(5): 226-30.
- Barbono, D.M., Ma, Y., Santos, M.V. Influence of raw milk quality on fluid milk shelf life. J. Dairy Sci., 2006; 89: E15-E19.
- Ruegg, P.L., Tabone, T.J. The relationship between antibiotic residue violations and somatic cell counts in Wisconsin dairy herds. J. Dairy Sci., 2000; 83: 2805-2809.
- Savelle, W.J., Wittum, T.E., Smith, K.L. Association between measures of milk quality and risk of violative antimicrobial residues in grade-A raw milk. J. Am. Vet. Med. Assoc., 2000; 217: 541-545.
- Jayarao, B.M., Henning, D.R. Prevalence of foodborne pathogens in bulk tank milk. J. Dairy Sci., 2001; 84: 2157-2162.
- Hogan, J. Human health risks associated with high SCC milk.. Proceedings of the 44th Annual Meeting on National Mastitis,. Jan 16-19, 2005, Orlando, FL.,. 2005; pp: 73-7
- Straley, B.A., Donaldson, S.C., Hedge, N.V., Sawant, A.A., Srinivasan, V., Oliver, S.P., Jayarao, B.M. Public health significance of antimicrobial- resistant gram-negative bacteria in raw bulk tank milk. Foodborne Pathog. Dis., 2006; 3: 222-233.
- Sorhaug, T., Stepaniak, L. Psychrotrophs and their enzymes in milk and dairy products quality aspects. Trends in Food Sci. Technol., 1997; 8: 35-41.
- Lindemann, P.C., Foshaugen, I., Lindemann, R. Characteristics of breast milk and serology of women donating breast milk to a milk bank. Arch. Dis. Childhood: Fetal and Neonatal Edition, 2004; 89(5): F 440-1.
- Rozolen, C.D., Goulart, A.L., Kopelman, B.I. Is breast milk collected at home suitable for raw consumption by neonates in Brazilian public neonatal intensive care units? J. Human Lactation, 2006; 22(4): 418-25.
- Carroll, L., Davies, D.P., Osman, M., McNeish, A.S. Bacteriological criteria for feeding raw breast-milk to babies on neonatal units. Lancet. 1979; 2(8145): 732-3.
- Karimi, M., Eslami, Z., Lotfi, M.H., Nori, S., Zandi, H., Taghipour-Zahir, S., Akhondzardaini, R. Bacterial contamination of expressed breast milk in neonatal intensive care unit. ZJRMS, 2013; 15(4): 48-52.
- Wallner, S., Schroeder, M., Kendall, P. Raw Milk: Why Pasteurize? 2006; http://www.ext.colostate.edu/safefood/newsltr/v10n2s04.html
- Grant, I.R., Pope, C.M., O’Riordan, L.M., Ball, H.J., Rowe, M.T. Improved detection of <I>Mycobacterium avium</I> subsp. paratuberculosis in milk by Immuomagnetic PCR. J. Vet. Microbiol. 2007; 77: 369-378.
- Orloff, S.L., Wallingford, J.C., McDougal, J.S. Inactivation of human immunodeficiency virus type 1 in human milk: Effects of intrinsic factors in human milk and pasteurization. J. Human Lactation, 2008; 9(1): 13-17.
- Egbere, O.J., Pam, K.V., Adesheyan, K.D., A’Kadir, T., Oyero, S.K. Effects of pasteurisation on survival patterns of microorganisms and vitamin C retention in kunun zaki. Afr. J. Biotechnol., 2009; 8(23): 6603-6607.
- Macaulay, D.M., Hawirko, R.Z., James, N. Effect of Pasteurization on Survival of Certain Psychrophilic Bacteria. J. Appl. Environ. Microbiol., 1963; 11(2): 90-9.
- Momoh, J.E., Udobi, C.E., Orukotan, A.A. Improving the Microbial Keeping Quality of Home Made Soymilk Using a Combination of Preservatives, Pasteurization and Refrigeration. Brit. J. Dairy Sci., 2011; 2(1): 1-4.
- Savage, W. G. The effect of pasteurization upon the organisms found in milk and its nutritive properties. Testing Huygens Principle, 1999; 221(5713): 429-431
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


