ISSN: 0973-7510
E-ISSN: 2581-690X
Hospital acquired infections caused by Non-fermentative gram-negative bacteria (NFGNB) have features that are of specific concern, detection of which is helpful for prevention and for better quality of healthcare in hospital premises. Present study was conducted to find out the nosocomial infections caused by NFGNB and its sensitivity pattern. Present microbiological study was carried out at C. U. Shah Medical College. Clinical samples which were included in this study were in accordance with inclusion criteria laid down in the research protocol, which were obtained during January 2018 to June 2020.Samples were collected and processed with the help of automated identification and antimicrobial susceptibility testing instrument i.e. Vitek – 2 as per standard guidelines. Statistical analysis was done by Microsoft Excel. Total 385 were classified as NFGNB from 1000 confirmed isolates causing Hospital acquired infection (HAI). Respiratory site infection was most common type of HAI i.e. 48%. While most common isolate was Pseudomonas sp.i.e. 42% followed by Acinetobacter baumannii i.e. 31%. Medicine ward showed maximum isolates i.e. 33%, followed by Surgery ward i.e. 23%. Maximum NFGNB isolates encountered from sputum sample i.e. 25% while blood shows only 6%. Trimethoprim/sulfamethoxazole and ciprofloxacinwere disclosedmaximum resistant. This study reports 385NFGNB isolates causing HAI. Such alarming resistance among NFGNB spreading the threat in medical care and epidemiology as they act as nosocomial pathogen. These isolated pathogens being the major cause of illness particularly in hospitalized patients.
Hospital acquired infection, Nosocomial infection, Non fermentative gram-negative bacteria, Antimicrobial susceptibility pattern
Hospital Associated Infections or nosocomial infections are those infections that are neither present nor incubating at the time the patient was admitted to the health care facility1. Non-fermentative gram-negative bacteria (NFGNB) are of specific concern as they are extremely competent to produce higher antibiotic drug resistance with the help of enzymes, especially in the presence of antibiotic selection pressure. These multidrug resistances produced by NFGNB are of higher clinical concerns which are increasingly being reported worldwide. These infections are most commonly associated with invasive devices or surgical procedures. However respiratory tract and bloodstream infections are the most dangerous HAI infections. Pseudomonas sp. and Acinetobacter sp. are most commonly encountered NFGNB associated with hospital-acquired infections. Unfortunately, these organisms are geared up with ESBL and a Carbapenemase enzyme which helps in the emergence of hospital acquired infections2.
NFGNB are being major cause of human illness due to act as a nosocomial pathogen. These isolates are resistant to many antibiotics. This resistance is due to genetic material which may be transferred between one bacterium to other. Hence it is important to report these isolates and their antibiotic resistance pattern routinely3.
Hospital acquired infections occurs in patients admitted in hospital or other clinical care facility. These infections are not only limited to patients, it also affects the health care workers including doctors. CDC suggests that 10% of all hospital infections are nosocomial infection. HAI caused by NFGNBs has been increasingly reported in the literature. Isolation of these NFGNB needs special biochemical reaction set or automated identification instrument like Vitek-2 which is time and resource consuming process4,5.
In developing countries like India, HAI surveillance studies especially by NFGNBs are not adequate. Efforts are made by ICMR (Indian Council of Medical Research) and have developed an Antimicrobial Resistance Surveillance Network. Five major hospitals of India are included in this network, however much more is still needed to be done for antimicrobial reservation and stewardship. This study was commenced with the objective of studying the nosocomial infections caused by NFGNB and its sensitivity pattern in a tertiary care teaching hospital of Gujarat. This baseline knowledge may deliver essential information to frame antibiotic policy in our hospital5.
This observational laboratory based study was approved by Institutional ethics committee and scientific review committee of the institute. Clinical samples which were included in this study were in accordance with inclusion criteria laid down in the research protocol. Study was carried out in January 2018 to June 2020. Samples were collected and processed with the help of Vitek – 2 automated identification and antimicrobial susceptibility testing instrument as per standard guidelines. Statistical analysis was done by Microsoft Excel1.6.
All the patients admitted in various wards or ICUs for more than 48 hours and developed the secondary infection other than present or incubating at the time of admission were included in the study. Also a detailed proforma was filled from the patients showing various clinical history, risk factors and demographical data of the patient. Hospital acquired infection was identified by with the help of detailed clinical history, active sign and symptoms and duly filled proforma.
Various clinical samples submitted to microbiology laboratory for Culture and Sensitivity testing from patients admitted in hospital. Specimens were primarily identified through gram stain and further cultured on MacConkey’s agar and blood agar; incubated at 35˚C, overnight. “BacT/Alert automation blood culture system” was used for blood and other body-fluid specimens6.
Identification of NFGNB was done by colonies having characteristic property of “non-lactose fermenter” and “non acidify butts”; on MacConkey’s and Triple sugar iron (TSI) respectively. Final identification and antimicrobial susceptibility testing were carried out by Vitek 2 automated system as per latest CLSI guidelines7,8.
Total 1000 patients were identified as Hospital acquired infection during January 2018 to June 2020, out of which 38.5% (385/1000) isolates were found as NFGNB. Maximum NFGNB strains of HAI were isolated from Respiratory site infection i.e. 48%. Out of 385 isolates 32% were from surgical site, 14% were from urinary tract and 6% were from blood stream.
Specimen wise distribution of isolates is shown in Table – 1. Maximum NFGNB isolates encountered from sputum sample i.e. 25% while blood shows only 6%.
Table (1):
Specimen wise distribution of isolates (n=385).
Specimens |
No. of isolates |
Percentage |
|---|---|---|
Sputum |
98 |
25% |
ET |
68 |
18% |
Swab |
65 |
17% |
Pus |
53 |
14% |
Urine |
53 |
14% |
Blood |
23 |
6% |
BAL |
21 |
5% |
Tissue |
04 |
1% |
– |
385 |
100% |
Maximum percentage of NFGNBs were from Medicine ward i.e. 33 % followed by surgery ward i.e.23%, MICU i.e.20%, Orthopedic ward i.e.9% and Respiratory ward i.e. 6 %.( Table – 2)
Table (2):
Department/ Unit wise distribution of isolates (n=385).
Department |
No. of isolates |
Percentage |
|---|---|---|
Gen. Medicine |
128 |
33% |
Surgery |
87 |
23% |
Intensive Care Unit |
77 |
20% |
Orthopedics |
35 |
09% |
Respiratory |
23 |
06% |
Pediatrics |
21 |
5% |
Otolaryngology |
06 |
2% |
Neonatal ICU |
04 |
1% |
Obstetrics and Gynecology |
04 |
1% |
Total |
385 |
100% |
In this study, the most common isolate was Pseudomonas sp., having 42% incidence, followed by Acinetobacter baumannii i.e.31% and Pseudomonas aeruginosa i.e.19 %. (Table – 3)
Table (3):
Organism wise distribution of isolates (n=385).
Organism |
No. of organisms |
Percentage |
|---|---|---|
Pseudomonas sp. |
164 |
42% |
Acinetobacter baumannii |
118 |
31% |
Pseudomonas aeruginosa |
74 |
19% |
Acinetobacter lwoffii |
10 |
3% |
Pseudomonas putida |
09 |
2% |
Sphingomonas paucimobilis |
07 |
2% |
Morganellamorganaii |
03 |
1% |
Total |
385 |
100% |
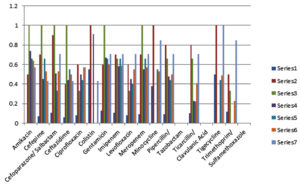
According to antibiotic susceptibility pattern of NFGNB isolates, Pseudomonas sp. noted maximum resistance against Trimethoprim- sulfamethoxazole i.e. 55% and maximum sensitivity against Amikacin i.e.64%, Gentamicin i.e.68% and Imipenem i.e.59%. Colistin showed 55 % sensitivity rates against Acinetobacter baumannii which was maximum. All other antibiotics gave less than fifty percent sensitivity against Acinetobacter baumannii isolates. (Fig. 1).
Fig. 1. Antibiotic sensitivity pattern of isolates.
Legend for Fig. 1. shows antibiotic sensitivity pattern of 385 isolates; X axis shows names of Antibiotics and Y axis shows sensitivity index from 0 to 1,maximum sensitivity 1 means 100% sensitive, Organisms in Series 1 Acinetobacter baumannii (n=118), Series 2 Acinetobacter lwoffii (n=10), Series 3 Morganella morganii (n=3), Series 4 Pseudomonas aeruginosa (n=74), Series 5 Pseudomonas putida (n=9), Series 6 Pseudomonas sp.(n=164), Series 7 Sphingomonas paucimobilis (n=7) (data on which this graph is prepared is given in appendix)
Hospitalized patients are more prone to nosocomial infections due to various risk factors like extreme age group, weak immunity and chronic diseases. Moreover highly resistant nosocomial pathogen in hospital environment and various invasive devices make them even more challenging to manage. Different nosocomial pathogens can get entry through indwelling urinary catheters, mechanical ventilators, intra vascular central lines, intra vascular cannulas and various medical procedures and causes hospital acquired infection. In our study respiratory tract infection was most common type of HAI. The most common respiratory tract infection found in our study was pneumonia caused by NFGNB. The frequent use of medical devices like mechanical ventilators, nebulizers, oxygen masks etc within different patients in hospital may be the one of the cause of Hospital acquired respiratory tract infection.
In present study 38.5% (385/1000) hospital acquired isolates were found as NFGNB. This data is higher than the studies of Sharma D et. al.3 and Agarwal S et. al.9 in which an isolation rate was 25% and 27%, respectively. A study by Moolchandani K et. al. from South India showed similar results of 38% NFGNB isolation5. These differences in the isolation rate of various NFGNB isolates in various health care settings are well expected as they depend on different infection control practices and other local variables.
Antibiotic resistance is real threat with NFGNB, including Pseudomonas and Acinetobacter, two nosocomial pathogens commonly isolated and reported in the literature9. Most common NFGNB isolated in the present study was Pseudomonas sp. in 42% and Acinetobacter baumannii in 31%, in the present study.
Maximum numbers of isolates were obtained from respiratory site i.e. 48%. Similar findings were also reported by Sharma D et. al.3 and Moolchandani K et. al.5 However high prevalence of hospital acquired surgery site infection has been reported about 28% in study of Sharma D et. al.3 which is almost similar with present study i.e. 32%.
In present study predominant isolates were Pseudomonas sp. i.e. 42%. It is higher than the study done in South India5 which shows 19% of Pseudomonas sp. isolates. However, Acinetobacter baumannii shows 29.5% in study of Sukumaran J et. al.10 which is similar to present study i.e. 31%. Many investigators also reported these alarming data of NFGNB in their studies of hospital acquired infections4.
NFGNB isolates showed high degree of resistance to almost all antibiotics including beta lactums, quinolones and sulfonamides. This data is supported by studies of Moolchandani K et.al.5 and Sharma D et. al.3 The most common drug resistant organism was Acinetobacter baumannii, which showed 89% resistance against Imipenem; and 92% against cephalosporin, which is a serious threat. Moolchandani et.al.5 had reported similar data in resistance pattern for Acinetobacter baumannii to various antibiotics. Colistin and Tigecycline were found most effective against Acinetobacter baumannii compared to other groups of antibiotics. It can serve as potential drug/s for the treatment against this isolate.
Resistance among these NFGNB isolates is important to know because they could become responsible for outbreaks of nosocomial infections11. They may be cause of increase in hospital costs as well as stay, morbidity and mortality12. Regular surveillance and record of these isolates may help us to control nosocomial infections and establish antibiotic policy of the hospital10-13. Concern for “Healthcare Associated Infections (HAIs)”, in Intensive Care units is also great concern which requires novel screening as well as laboratory diagnostic methods14. 20 percent of isolates of this study were from specimens obtained from MICU. Detecting sensitivity and resistance pattern helps clinicians in treating and controlling HAIs in critically ill patients. Hence, early detection of the same is important in controlling HAIs.
Limitation
Limitation of our study was that we have not analyzed the therapeutic outcome of patient with hospital acquired infection caused by NFGNB organisms. Second, the study does not include occupational hospital acquired infections among staff of the hospital.
The present study on the NFGNB and its antimicrobial susceptibility shows 385 isolates with HAI. Respiratory site infection was most common type of HAI. The most common isolates among NFGNB were Pseudomonas sp. and Acinetobacter baumannii. Medicine ward was most common location for isolating NFGNB causing HAI. Over all, NFGNB showed high resistance rates against Trimethoprim /sulfamethoxazole and least resistance against Amikacin.
To conclude, the percentage of multidrug resistance NFGNB causing HAI was high in our institutions which is not only alarming but enforces us for infection control measures which are efficient and sustained. These measures include hospital surveillances, hand washing practices, and periodic training of the hospital staff.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All the listed authors have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
The study was approved by the institutional ethics committee (Human Research), C.U. Shah Medical College, Gujarat, India.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Ducel G, Fabry J, Nicolle L. Prevention of Hospital Acquired Infections. A practical guide. 2nd ed. WHO manual. 2012.
- Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362(19):1804-1813.
Crossref - Sharma D, Vyas N, Sinha P, Mathur A. Non fermentative gram negative bacilli as nosocomial pathogens: Identification and antibiotic sensitivity in clinical samples of indoor patients. Nepal Journal of Medical Sciences. 2014;3(2):101-105.
Crossref - Panjwani DM, Lakhani SJ, Mehta SJ, Kikani KM, Madaan K. A Study of Hospital Acquired Bacterial Infections and its Antimicrobial Susceptibility Pattern in a Teaching Hospital of Gujarat, India. Int J Curr Microbiol Appl Sci. 2020;9(02):1399-1408.
Crossref - Moolchandani K, Satry A, Deepashree R, Sistla S, Harish B, Mandal J. Antimicrobial Resistance Surveillance among Intensive Care Units of a Tertiary Care Hospital in Southern India. J Clin Diagn Res. 2017;11(2):DC01-DC07.
Crossref - Forbes BA, Sahm DF, Weissfeld AS, edts; Bailey and Scott’s. Diagnostic microbiology. 12th ed. Mosby Elsevier. 2007.
- Winn W Jr, Allen S, Janda W, Koneman E, Procop G, Schreckenberger P, et al editors. Nonfermenting gram negative bacilli. In: Koneman’s color atlas and textbook of Diagnostic Microbiology. 6th ed. USA: Lippincott Williams and Wilkins Company. 2006;305-391.
- Weinstein M. Performance standards for antimicrobial susceptibility testing. M100. 28th ed.: Clinical and Laboratory Standards Institute. 2018.
- Agarwal S, Kakati B, Khanduri S, Gupta S. Emergence of Carbapenem Resistant Non-Fermenting Gram-Negative Bacilli Isolated in an ICU of a Tertiary Care Hospital. J Clin Diagn Res. 2017;11(1):DC04-DC07.
Crossref - Sukumaran J, Sriram L, Sumathi G. Nonfermentative gram negative bacilli- characterisation and antibiotic resistant pattern study from a tertiary care hospital. Indian Journal of Basic and Applied Medical Research. 2014;3(4):227-232.
- Chawla K, Vishwanath S, Munim FC. Nonfermenting Gram-negative Bacilli other than Pseudomonas aeruginosa and Acinetobacter Spp. Causing Respiratory Tract Infections in a Tertiary Care Center. J Glob Infect Dis. 2013;5(4):144-148.
Crossref - Barrasa-Villar JI, Aibar-Remon C, Prieto-Andres P, Mareca-Donate R, Moliner-Lahoz J. Impact on Morbidity, Mortality, and Length of Stay of Hospital-Acquired Infections by Resistant Microorganisms. Clin Infect Dis. 2017;65(4):644-652.
Crossref - Mehta Y, Gupta A, Todi S, et al. Guidelines for prevention of hospital acquired infections. Indian J Crit Care Med. 2014;18(3):149-163.
Crossref - Chaturvedi A, Banashankari GS. Utility of a novel chromogenic medium as a screening method in the detection of carbapenemase producing Enterobacteriaceae. J Lab Physicians. 2017;9(3):202-206.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.