ISSN: 0973-7510
E-ISSN: 2581-690X
Skin and soft tissue infections (SSTIs) are the most common type of infection globally and also in India. Periodic evaluation of data on microbial profiling and antibiogram trend analysis is mandatory for creating a timely empirical treatment guideline and a proper antimicrobial cycling plan. In the current study, retrospective analysis of the data from 3,570 samples collected from suspected SSTI over one year was performed. Analysis was done on the clinical condition, causative agent/s identified and their antimicrobial susceptibility according to the standard guidelines. Seventy-three percent of samples yielded positive growth, with majority being unimicrobial infections. Gram-negative bacteria (GNB) were more commonly associated with infections from in-patients while Gram-positive cocci (GPC) were seen among out-patients. More than 70% of isolates among GNB were found susceptible to Carbapenems, piperacillin-tazobactam, amikacin and chloramphenicol. Among GPC, maximum sensitivity was seen to glycopeptides, linezolid followed by chloramphenicol, tetracycline, clindamycin and amikacin. Unusual resistance patterns like Penicillin resistant Ampicillin sensitive Enterococcus faecalis (PRASEF)strains, Multidrug resistance (MDR) and Extended drug resistance (XDR) were higher among Enterococci spp, MRSA, Acinetobacter spp and Klebsiella spp. Resistance to penicillin and cephalosporin drugs were high among GNB and GPC. Carbapenems, aminoglycosides, glycopeptides and broad-spectrum antibiotics are found sensitive and potential choices for empirical therapy. As prevalence of MDR and XDR strains were high and the trend analysis shows likelihood of few of the first and second line drugs becoming sensitive in future, we conclude that continued analysis of bacterial profiling and AMR pattern analysis among SSTI is essential.
SSTI, Antibiogram pattern analysis, South India, bacterial profile, PRASEF
Skin and soft tissue infections (SSTIs) is the most common type of infection globally as well as in India. It constitutes around 29-32% of all infections in 2018-20191,2. It can range from a simple superficial lesion that can resolve from topical applications and oral antibiotics, to serious conditions requiring surgical interventions and parenteral therapy. Common uncomplicated infections are furuncle, carbuncle, impetigo, secondary infections in a wound, superficial ulcers, etc. The lesion is said to be complicated when there is involvement of subcutaneous plane and adjacent soft tissues as seen in necrotizing fasciitis, extensive cellulitis and deep-seated ulcers. In addition, patient’s comorbid conditions like uncontrolled diabetes mellitus, ischemia, immunocompromised state add on to the progress of the lesion and failure in treatment3-5. These infections are caused by a wide range of bacteria and fungi. Most common organisms involved in SSTI are Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa and Candida spp1,5.
Most of these uncomplicated infections are treated in outpatient departments (OPDs) empirically while awaiting culture and sensitivity results. In smaller outpatient settings diagnosis and treatment is based largely on clinical decision making and the established epidemiological data published from time to time6. Study of the bacterial profile causing various types of infection guides us towards the correct choice of antibiotic. Antibiogram pattern of the isolates are also known to change periodically depending the prescription patterns, due to selection pressure on the strains causing infections. In the era of growing antimicrobial resistance, a functional antimicrobial stewardship program is essential in every tertiary care hospital. Periodic evaluation of data on bacterial profiling and antibiogram trend analysis is mandatory creating a proper antimicrobial cycling plan. This helps in avoiding treatment failures and selection of resistant strains due to mistreatment6-8.
The inadvertent use of antibiotics leads to emergence of drug resistant pathogens, leading to poor prognosis9,10. A predictable bacterial profile of the wound infections is very important for clinicians, to start empirical treatment. The purpose of this study is to show the spectrum of aerobic bacterial and fungal profile in relation to different types of skin and soft tissues infections over a year period and its periodic sensitivity pattern, analyzing the trends.
The study was conducted in the Department of Microbiology at SRM Medical college hospital and research center, Kattankulathur, Tamilnadu after obtaining Institution’s permission and Institutional Ethical clearance. Data (except patient identification details)of all the samples sent to Diagnostic Microbiology laboratory for aerobic culture and sensitivity, from January 2019 to December 2019 were collected along with the provisional diagnosis mentioned during testing. All purulent samples, swabs from wound and ulcers, tissue biopsy materials, granules from the SSTI lesions, processed and reported, were studied. Identification of isolates was performed using manual biochemical method and antimicrobial susceptibility testing was performed by Kirby Bauer disk diffusion method and E-strip method, in our laboratoryand interpreted as per Clinical Laboratory Standard Institution (CLSI) guidelines M100 S30 and M44-A211,12 and manufacturer’s instruction. Data on date of sampling, type of sample, sample source location, provisional clinical diagnosis, organism isolated from the sample, diameters of zone of inhibition for the individual isolates and special resistance patterns were computed in Microsoft excel sheets and analyzed.The zone of inhibition of individual drugs against each species isolated were analyzed for trend of increase, decrease or nil change in their diameters.
A total of 3,570 samples were processed for SSTI during the study period. Table 1 shows the overall distribution of positive sample with respect to patient clinical condition, unimicrobial or polymicrobial status. A total of 2662 isolates were analyzed for the antibiogram pattern. Isolates analyzed were from OPD (9.8%), wards (81%) and ICU (9.2%).
Table (1):
Distribution of samples analyzed in the study based on type of growth and clinical diagnosis.
| Total samples analyzed 3570 |
Growth positive samples 2602 (72.9%) |
Unimicrobial infections 2542 (97.7%) |
SSTI ulcers | Diabetic foot ulcers | Superficial and deep abscess | Non healing ulcers | Cellulitis | Surgical site infections | Diagnosis Unknown |
|---|---|---|---|---|---|---|---|---|---|
| Polymicrobial infections 60(2.3%) |
1793 (67.37%) | 465 (17.47%) | 136 (5.11%) | 27 (1.01%) | 107 (4.02%) | 83 (3.12%) | 51 (1.9%) |
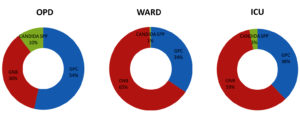
Fig. 2. Percentage distribution of Gram-positive cocci, Gram-negative bacilli and Candida spp isolates among samples from OPD, Ward and ICU
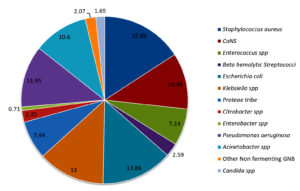
Distribution of bacterial profile remained constant throughout the year with mild variations. Fig. 1 shows the prevalence of various pathogens causing SSTI in our study population. The prevalence of gram-positive and gram-negative bacterial infections were 36.1% and 62.2 % respectively. Prevalence of infections with Candida spp was 1.69 percent. Distribution of Gram-positive bacteria, Gram-negative bacteria and Candida spp. from OPD, wards and ICU are shown in Fig. 2.
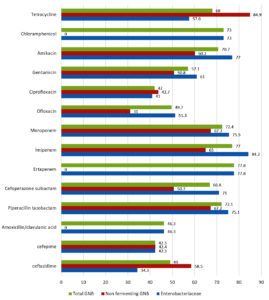
Fig. 3. Overall analysis and split analysis (Non-fermenting GNB and Enterobacteriaceae) of antibiotic susceptibility pattern of Gram-negative bacilli causing SSTI. Percentage of susceptible strains to individual antibiotics tested is shown.
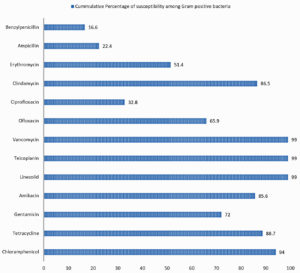
Fig. 4. Cumulative percentage of susceptibility of Gram-positive bacteria causing SSTI to individual antibiotics tested.
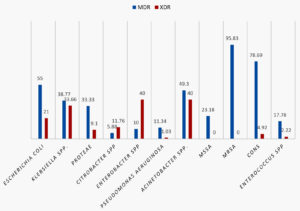
Fig. 5. Percentage prevalence of Multidrug resistant (MDR) and extended drug resistant (EDR) strains among various organisms studied
Antibiotic susceptibility patterns of individual groups of organism isolated are shown in the Tables 2, 3 and 4. Difference observed in susceptibility pattern of fermenting and non-fermenting GNB in comparison to all GNBs isolated is shown in Fig. 3. Cumulative percentage of susceptibility among gram-positive bacteria is shown in Fig. 4. Prevalence of MDR and XDR strains among the isolates is shown in Fig. 5.
Table (2):
Antibiotic susceptible pattern of Gram-positive cocci analyzed in the study.
Antimicrobials |
Staphylococcus aureus [n=422] Sensitivity % |
Coagulase negative Staphylococcus spp.[n=289] Sensitivity % |
Enterococcus spp. [n=190] Sensitivity % |
Beta hemolytic Streptococcus spp. [n=69] Sensitivity % |
|---|---|---|---|---|
Benzyl penicillin |
4.7 |
6.1 |
49.2 |
39.6 |
Ampicillin |
4.7* |
6.1* |
88.2 |
96.1 |
CefoxitinϮ |
62 |
38.4 |
IR |
– |
Erythromycin |
59.4 |
47.4 |
36.5 |
50 |
Clindamycin € |
90.1 |
82.1 |
IR |
86.4 |
Ciprofloxacin |
31.8 |
41.1 |
5.5 |
– |
Levofloxacin £ |
– |
– |
– |
71.4 |
Ofloxacin |
69.9 |
60.2 |
– |
66 |
Tetracycline |
90.6 |
89.5 |
– |
84.1 |
Vancomycin |
– |
– |
98.9 |
100 |
Teicoplanin |
– |
– |
98.9 |
– |
Linezolid |
100 |
98.2 |
98.4 |
97 |
Trimethoprim/ Sulphamethoxazole |
90.1 |
65.1 |
IR |
54.9 |
Gentamicin [30µg] |
80 |
61.9 |
IR |
– |
Gentamicin [120µg] |
– |
– |
72.2 |
– |
Amikacin |
95.3 |
74.8 |
IR |
– |
Chloramphenicol |
96.6 |
95.6 |
IR |
87.8 |
IR- Intrinsic Resistance
*Sensitivity derived based on results of Benzyl penicillin
Ϯ Cefoxitin screen was used to detect susceptibility to Methicillin.
€ Results analyzed based on zone of inhibition and D test results.
£ Number of isolates studied were too low for extrapolating the results to study population
Table (3):
Antibiotic susceptible pattern of Gram-negative bacilli analyzed in the study.
Antimicrobials |
Escherichia coli [n=368] Sensitivity % |
Klebsiella spp. [n=347] Sensitivity % |
Proteae tribe [Proteus spp, Morganellaspp, [n=198] Providencia spp. ] Sensitivity % |
Citrobacter spp. [n=60] Sensitivity % |
Enterobacter spp. [n=19] Sensitivity % |
Acinetobacter spp [n=282] Sensitivity % |
Pseusomonas aeruginosa [n=318] Sensitivity % |
|---|---|---|---|---|---|---|---|
Ampicillin |
13.7 |
IR |
40 |
IR |
IR |
IR |
IR |
Cefazolin |
18.6 |
18.4 |
48.3 |
– |
IR |
IR |
IR |
Cefuroxime |
23.3 |
22.8 |
10.3 |
– |
IR |
IR |
IR |
Ceftriaxone |
24.7 |
28.3 |
52.1 |
51.7 |
IR |
13.3 |
IR |
Cefotaxime |
23.8 |
23 |
48.6 |
– |
IR |
– |
IR |
Ceftazidime |
34.5 |
28.6 |
43.7 |
– |
IR |
10.6 |
83.7 |
Cefepime |
31.3 |
36.3 |
64.4 |
71.7 |
IR |
22.2 |
98 |
Amoxicillin clavulanate |
39.2 |
– |
77.9 |
42.6 |
IR |
IR |
IR |
Piperacillin tazobactam |
76.2 |
58 |
99.5 |
86.7 |
60 |
31.8 |
98.1 |
Ertapenem |
81.4 |
61.3 |
97.4 |
88.3 |
60 |
IR |
IR |
Imipenem |
85.1 |
76.0 |
98 |
88.3 |
60 |
32.1 |
93.3 |
Meropenem |
84.8 |
51.3 |
98 |
88.3 |
60 |
41.5 |
90.5 |
Gentamicin |
65.2 |
52.7 |
68.4 |
61.9 |
45 |
22.9 |
78 |
Amikacin |
87.9 |
65.6 |
74.6 |
84.7 |
70 |
26 |
92 |
Ciprofloxacin |
24.6 |
51.9 |
50.6 |
68 |
35 |
16.3 |
69.7 |
Ofloxacin |
39.5 |
59.1 |
– |
81.1 |
– |
31 |
– |
Trimethoprim/ Sulphamethoxazole |
36.9 |
45.4 |
– |
69.2 |
25 |
30 |
IR |
Chloramphenicol |
70.3 |
74.4 |
74.6 |
– |
– |
IR |
IR |
Tetracycline |
39.3 |
72.9 |
IR |
– |
72.7 |
84.9 |
IR |
IR- Intrinsic Resistance
Table (4):
Percentage of susceptible strains among Candida species analyzed in the study.
Antifungals |
Candida albicans n=23 [%] |
Non albicans Candida spp n=22 [%] |
Total Candida spp n=45[%] |
|---|---|---|---|
Fluconazole |
21 [91.3] |
17 [77.3] |
38 [84.4] |
Voriconazole |
21 [91.3] |
19 [86.4] |
40 [88.9] |
Itraconazole |
17 [73.9] |
11 [50] |
28 [62.2] |
Amphotericin B |
15 [65.2] |
13 [59.1] |
28 [62.2] |
On analyzing the trends in zone sizes, among the Gram-positive cocci (GPC), a gradual increase in diameters of zone of inhibition, over a period of one year, was observed with Erythromycin, clindamycin, Benzyl Penicillin and Ampicillin. Cefoxitin screen showed an increase in susceptibility for Staphylococcus aureus but decreasing trend for Coagulase negative Staphylococcus species (CoNS) isolates. Increasing trend was observed for High level Gentamicin against Enterococcus spp.
Among the Enterobacteriaceae family, there was a generalized increasing trend (with zone diameters) observed with ciprofloxacin, ofloxacin, cotrimoxazole and cefepime. Gentamicin showed an increasing trend with Escherichia coli and Pseudomonas aeruginosa. Ampicillin and amoxicillin clavulanic acid showed an increasing trend with Proteus spp and Citrobacter spp.
In the current study 72.9% samples tested yielded growth. The prevalence of polymicrobial infections were low compared to other data available. Past studies done in India showed that infections due to Gram-positive bacteria is predominant in case of SSTIs13,14.Recent studies from western world have reported GPC as the most predominant organism accounting for more than 50% of the SSTIs. Among the Gram-positive bacteria, Staphylococcus aureus in particular, is the most predominant bacteria causing SSTI in other countries15-18. However recent studies in India have shown a rising trend with gram-negative bacterial infections in this disease category. Also, our current study shows that prevalence of Gram-negative bacteria is twice as high as gram-positive bacteria. Similar results were projected in annual report of Indian Council of Medical Research for year 2018 and 2019 on antimicrobial resistance surveillance throughout the country1,2.Prevalence of infections due to Gram-positive cocci (54%) and candida (10%) was higher in OPD patients (community). Prevalence of infections due to Gram-negative bacilli (GNB) was 59-65%, which was higher among ICU and in-patient category. There is an increase in incidence of Gram-negative bacterial and fungal infections in recent days19,20.Given the increase in multidrug resistance among Gram-negative bacteria, it has become difficult to treat the SSTIs due to change in prevalence and resistance trends. In the current study, prevalence of Candida infection was 1.9% which is slightly lower than the national data (2.5%)1.
When analyzed individually Staphylococcus aureus (15.85%) followed by Escherichia coli (13.86%), Klebsiella spp (13%) and Pseudomonas aeruginosa(11.95%) were the leading causes of SSTI in our study population. Coagulase negative Staphylococcus spp(CoNS)were isolated in significant colony counts from 10.86% of cases. Their prevalence is increasing over past few decades1,2,13,20,21. It was found more often associated with Diabetic foot ulcers, implant related infections and wound infections. Approximately 30% of the CoNS isolated were Staphylococcus hemolyticus. Acinetobacter spp which was once associated exclusively with nosocomial infections like ventilator associated pneumonia is now seen associated with community acquired infection22. Prevalence of Acinetobacter spp was 10.6% in the current study. Similar results were observed in national and some of the regional studies1,2,23.Fifty-five (2.07%) isolates were non fermenting gram-negative bacilli. Of the 55, 12 isolates were Stenotrophomonas maltophila, 4 were Burkholderia pseudomallei and 6 were Sphingomonas paucimobilis and 33 isolates were not identified further.
It is important to monitor the changing trends in bacterial and fungal infection and their antimicrobial susceptibility pattern overtime, to provide adequate antimicrobial therapy for controlling infection and preventing morbidity. Prevalence of methicillin resistance was 62.02% and 38.37% among Staphylococcus aureus and CoNS respectively, adding up to 50.2 % among Staphylococcal infections. All Staphylococcus spp were sensitive to Vancomycin, Teicoplanin and Linezolid. Antimicrobials showing 90% and above susceptibility against Staphylococcus spp were Chloramphenicol, Tetracycline, Trimethoprim/Sulphamethoxazole and Clindamycin. Gentamicin and amikacin showed 80% and 95.3% susceptibility in Staphylococcus aureus. The susceptibility rates were lower among CoNS.
Susceptibility to vancomycin, teicoplanin and linezolid was 98.3-98.9% among Enterococcus spp. Ampicillin and high-level aminoglycosides were susceptible in 88% and 72% of the isolates. Susceptibility to Benzyl penicillin was 49.2 percent. Among these isolates 38.8% were penicillin resistant but ampicillin sensitive. Studies show that this may be due to polymorphism in pbp4gene in PRASEF strains1,2,24-26. Studies have shown that PRASEF isolates require E testing or broth dilutions determining their MICs, as the appropriate method to analyze susceptibility to piperacillin and imipenem. Disk diffusion cannot be considered as a reliable method of testing in these isolates27. However only one out of seventy isolates tested for imipenem showed unusual pattern of penicillin and imipenem resistance but ampicillin susceptible. Similar pattern was observed and studied by Metzidie et al.26 Remaining isolates had a well correlating results between penicillin, ampicillin, and imipenem.
Among the 69 Beta haemolytic streptococcus isolated, 40.5% was Streptococcus pyogenes, 23.2% was Streptococcus agalactiae, 23.2% was Group G or Group F Streptococci and 13% was non typable. Three fourth of the Group G or F strains were penicillin resistant. Majority of these strains were multidrug resistant (macrolide and fluoroquinolone resistance observed). All the strains were susceptible to Vancomycin, linezolid, and chloramphenicol. Other beta hemolytic Streptococcal isolates were susceptible to beta lactams and other group of antibiotics tested.
In the present study, the most common organism among Gram-negative bacteria was Enterobacteriaceae (37.26%) compared to non-fermenters (24.62%). Rate of Extended spectrum beta-lactamase (ESBL) producers was 40% (by combination disk test of cephalosporin/clavulanic acid) and Carbapenem resistance was 21.43% among Enterobacteriaceae. There were 277 (27%)isolates having a zone of inhibition of 6mm to third generation cephalosporins tested (ceftazidime, cefotaxime, ceftriaxone), ceftazidime clavulanic acid and cefoxitin. Percentage sensitive to ceftriaxone, cefotaxime and cefepime were 29.3%,33.3% and 42.3% respectively. Imipenem had a higher percentage of susceptibility among Escherichia coli(85%), Klebsiella spp(76%) and Pseudomonas spp(93%) compared to ertapenem and meropenem. Carbapenem resistance was highest in Acinetobacter spp(60-68%) followed by Klebsiella spp(40-50%) and Enterobacter spp(40%).
Gram-negative bacteria were observed to have less than 40% susceptibility to the following antibiotics – Ampicillin, cefazolin, cefuroxime, ceftriaxone, cefotaxime and trimethoprim / sulphamethoxazole. Susceptibility ranging from 40% to 50% was observed with cefepime, amoxicillin clavulanic acid, ciprofloxacin and ofloxacin.
Enterobacteriaceae isolated showed high susceptibility to Imipenem (84.2%), Ertapenem (77.8%), Amikacin (77%), Meropenem (75.5%), piperacillin-tazobactam (75.1%), chloramphenicol (73%) and cefoperazone sulbactam (71%). Non fermenting GNB (Pseudomonas spp and Acinetobacter spp) showed high susceptibility to tetracycline (84.9%), Meropenem (67.3%), piperacillin-tazobactam (67.2%), Imipenem (64.9%) and amikacin (60.2%). There is a decrease in susceptibility observed with beta lactam antibiotics compared to data from similar studies in previous years20,21,28,29
Among the Gram-negative bacilli isolated the overall resistance rates were highest with Acinetobacter spp followed by Klebsiella spp. Similar results were observed with studies from various parts of India21,30-32. Multidrug resistance(MDR)and extensive drug resistance (XDR)were seen in 38.1% and 14.88% of isolates respectively. Similar results were observed in studies from other parts of the country33-36. Majority of MDR was observed with Staphylococcus spp., Escherichia coli and Acinetobacter spp. XDR was highest among Acinetobacter spp, Klebsiella spp and Enterobacter spp Early detection and close monitoring of MDR and XDR bacterial strains is mandatory to reduce the menace of antimicrobial resistance which is now a global problem.
GPC were found to be more sensitive to tetracycline (88.7%) and chloramphenicol (94%) compared to GNB where sensitivity was 68% and 73% respectively.
Relatively high resistance to third generation cephalosporins with or without Betalactamase inhibitors was observed in the current study. This may be due to a large portion of the isolates being analyzed from ICU and patients admitted in wards.
Candida infection was found among patients with Diabetic foot ulcer (89%) predominantly in this study. Candida albicans and other non albicans species were equally prevalent in our study population. These isolates were most susceptible to Voriconazole (88.9%) followed by Fluconazole (84.4%) Currently fluconazole is used safely as oral antifungal among our population. Susceptibility data supports the continuation of the same.
Knowledge of the common causative agents of SSTIs and their antimicrobial susceptibility pattern is essential for the judicious administration of empirical treatment. Antimicrobial susceptibility of microorganisms varies with time and place. Hence a frequent data analysis and a good antibiotic stewardship must be enforced from primary, secondary, and tertiary care facilities.
In the current study, Carbapenem, Amikacin and Piperacillin-tazobactam were found to be the most sensitive antibiotics against GNBs. Vancomycin, Teicoplanin, Linezolid and clindamycin were found to be the most sensitive antibiotics against GPCs. Both GNB and GPC were found to be most sensitive to broad spectrum antibiotics tested- Tetracycline and chloramphenicol. Though antibiotics like fluoroquinolones, cotrimoxazoles, cefepime erythromycin, penicillin group of drugs were recorded to have higher percentage of resistance, there was a gradual increasing trend in zone diameters, indicating the possibility of gaining higher susceptibility percentage in due time.
ACKNOWLEDGMENTS
We would like to thank Head of microbiology and management for their support. We would like to thank our clinical team and laboratory’s technical staff for their support throughout the year.
CONFLICT OF INTEREST
All the authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
The study was approved by the institutional ethics committee, SRM Medical Hospital and Research Centre, Approval number 2018/IEC/2020.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Walia K, Ohri MGV, Sahni AK. Annual Report Antimicrobial Resistance Surveillance and Research Network January 2019 to December 2019.
- Walia K, Ohri MGV, Singh H, Madhumathi J, Kaur J. Annual Report Antimicrobial Resistance Surveillance and Research Network January 2018 to December 2018.
- Napolitano LM. Severe Soft Tissue Infections. Infect Dis Clin North Am. 2009;23(3):571-591.
Crossref - Dryden MS. Complicated skin and soft tissue infection. J Antimicrob Chemother. 2010;65(SUPPL. 3):35-44.
Crossref - Dryden MS. Skin and soft tissue infection: microbiology and epidemiology. Int J Antimicrob Agents. 2009;34:S2-S7.
Crossref - Fridkin SK. Routine cycling of antimicrobial agents as an infection-control measure. Clin Infect Dis. 2003;36(11):1438-1444.
Crossref - Beardmore RE, Pena-Miller R, Gori F, Iredell J, Barlow M. Antibiotic cycling and antibiotic mixing: Which one best mitigates antibiotic resistance? Mol Biol Evol. 2017;34(4):802-817.
Crossref - Van Duijn PJ, Verbrugghe W, Jorens PG, et al. The effects of antibiotic cycling and mixing on antibiotic resistance in intensive care units: a cluster-randomised crossover trial. Lancet Infect Dis. 2018;18(4):401-409.
Crossref - Kaur I. Analysis of Microbial Resistance & Prescription Preferences Using Antibiograms. Pharm Pharmacol Int J. 2016;4(7):502-508.
Crossref - Burnham JP, Kirby JP, Kollef MH. Diagnosis and management of skin and soft tissue infections in the intensive care unit: a review. Intensive Care Med. 2016;42(12):1899-1911.
Crossref - Clinical and Laboratory Standards Institute. CLSI M100 30th Edition. Vol. 30th, Journal of Services Marketing. 2020.
- CLSI. Method for Antifungal Disk Diffusion Susceptibility Testing of Yeasts; Approved Guideline- Second Edition. CLSI document M44-A2.Wayne, PA: Clinical and Laboratory Standards Institute; 2009.
- Thangavel S, Maniyan G, Vijya S, Venkateswaran C. A study on aerobic bacteriological profile and antimicrobial susceptibility pattern of isolates from pus samples in a tertiary care hospital. Int J Bioassays. 2017;6(03):5317.
Crossref - Afroz Z, Metri BC, Jyothi P. Bacteriological profile and antimicrobial susceptibility pattern of skin and soft tissue infections among gram negative bacilli in a tertiary care hospital of south India. J Pharm Sci Res. 2015;7(7):397-400.
- Ki V, Rotstein C. Bacterial skin and soft tissue infections in adults: A review of their epidemiology, pathogenesis, diagnosis, treatment and site of care. Can J Infect Dis Med Microbiol. 2008;19(2):173-184.
Crossref - Kaye KS, Petty LA, Shorr AF, Zilberberg MD. Current epidemiology, etiology, and burden of acute skin infections in the United States. Clin Infect Dis. 2019;68(Suppl 3):S193-S199.
Crossref - Ray GT, Suaya JA, Baxter R. Incidence, microbiology, and patient characteristics of skin and soft-tissue infections in a U.S. population: A retrospective population-based study. BMC Infect Dis. 2013;13:252.
Crossref - Fritz SA, Shapiro DJ, Hersh AL. National Trends in Incidence of Purulent Skin and Soft Tissue Infections in Patients Presenting to Ambulatory and Emergency Department Settings, 2000-2015. Clin Infect Dis. 2020;70(12):2715-8.
Crossref - Sultana S, Mawla N, Kawser S, Akhtar N, Ali MK. Current Microbial Isolates from Wound Swab and Their Susceptibility Pattern in a Private Medical College Hospital in Dhaka city. Delta Med Coll J. 2015;3(1):25-30.
Crossref - Biradar A, Farooqui F, Prakash R, Khaqri SY, Itagi I. Aerobic bacteriological profile with antibiogram of pus isolates. Indian J Microbiol Res. 2016;3(3):245-249.
Crossref - Singh A. A Prospective Study on Bacteriological Profile of Skin and Soft Tissue Infections with It’s Antimicrobial Sensitivity Pattern in a Tertiary Care Hospital. Int Arch BioMed Clin Res. 2019;5(1):71-73.
- Sebeny PJ, Riddle MS, Petersen K. Acinetobacter baumannii skin and soft-tissue infection associated with war trauma. Clin Infect Dis. 2008;47(4):444-449.
Crossref - Negi V, Pal S, Juyal D, Sharma MK, Sharma N. Bacteriological profile of surgical site infections and their antibiogram: A study from resource constrained rural setting of Uttarakhand state, India. J Clin Diagnostic Res. 2015;9(10):DC17-D20.
Crossref - Infante VHP, Conceicao N, De Oliveira AG, Da Costa Darini AL. Evaluation of polymorphisms in pbp4 gene and genetic diversity in penicillin-resistant, ampicillin-susceptible Enterococcus faecalis from hospitals in different states in Brazil. FEMS Microbiol Lett. 2016;363(7):1-5.
Crossref - Tan YE, Ng LSY, Tan TY. Evaluation of Enterococcus faecalis clinical isolates with penicillin-resistant, ampicillin-susceptible phenotype as reported by Vitek-2 Compact system. Pathology. 2014;46(6):544-550.
Crossref - Metzidie E, Manolis EN, Pournaras S, Sofianou D, Tsakris A. Spread of an unusual penicillin- and imipenem-resistant but ampicillin-susceptible phenotype among Enterococcus faecalis clinical isolates. J Antimicrob Chemother. 2006;57(1):158-160.
Crossref - Conceicao N, Rodrigues WF, De Oliveira KLP, et al. Beta-lactams susceptibility testing of penicillin-resistant, ampicillin-susceptible Enterococcus faecalis isolates: A comparative assessment of Etest and disk diffusion methods against broth dilution. Ann Clin Microbiol Antimicrob. 2020;19:43.
Crossref - Abhilash KP, Varghese S. Profile and outcome of patients presenting with skin and soft-tissue infections to the emergency department. Curr Med Issues. 2019;17(2):30-33.
Crossref - Singh B, Singh S, Khichy S, Ghatge A. Clinical Presentation of Soft-tissue Infections and its Management: A Study of 100 Cases. Niger J Surg. 2017;23(2):86-91.
Crossref - Effah CY, Sun T, Liu S, Wu Y. Klebsiella pneumoniae: An increasing threat to public health. Ann Clin Microbiol Antimicrob. 2020;19(1):1-9.
Crossref - Garbati MA, Godhair AIA. The growing resistance of klebsiella pneumoniae; The need to expand our antibiogram: Case report and review of the literature. African J Infect Dis. 2013;7(1):8-10.
Crossref - Manchanda V, Sanchaita S, Singh N. Multidrug resistant acinetobacter. J Glob Infect Dis. 2010;2(3):291-304.
Crossref - Basak S, Singh P, Rajurkar M. Multidrug Resistant and Extensively Drug Resistant Bacteria: A Study. J Pathog. 2016;2016:4065603.
Crossref - Leski TA, Taitt CR, Bangura U, et al. High prevalence of multidrug resistant Enterobacteriaceae isolated from outpatient urine samples but not the hospital environment in Bo, Sierra Leone. BMC Infect Dis. 2016;16:167.
Crossref - Bassetti M, Pecori D, Sibani M, Corcione S, De Rosa FG. Epidemiology and Treatment of MDR Enterobacteriaceae. Curr Treat Options Infect Dis. 2015;7(4):291-316.
Crossref - Logan LK, Weinstein RA. The epidemiology of Carbapenem-resistant enterobacteriaceae: The impact and evolution of a global menace. J Infect Dis. 2017;215(Suppl 1):S28-36.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.