ISSN: 0973-7510
E-ISSN: 2581-690X
This study reported the application of a next generation sequencing (NGS) analysis of yeast diversity in native Indonesian fruit, Durio kutejensis, collected from Borneo, Central Kalimantan. The analysis was designed to observe the microbial consortium associated with solid state fermentation (SSF) for amylase production. Together with the additional data from culture-dependent analysis, we observed the morphological features, molecular characteristics, and amylase concentration produced by each isolate. We performed Solid State Fermentation (SSF) for amylase production and the enzyme activity was then determined using UV-Vis spectrophotometer at 540 nm. Result obtained from metagenomic approach consist of 4 group that fungal species included in the Ascomycota identified as Botryosphaeria dothidea (1.35%), Lasiodiplodia crassispora (17.62%), Aureobasidium pullulans (55.02%), Paraphoma chrysanthemicola (11.38%), Preussia funiculate (1.90%), Sporormiella intermedia (0.82%), Myrothecium gramineum (1.35%), Fusarium oxysporum (6.24%), Fusarium proliferatum (3.25%) and Phialemoniopsis curvata (1.08%). The results of isolation using culturable medium in the form of YMA obtained 40 yeast isolates. A total of 40 representative isolates from durian fruit were screened, two positive amylase isolates based on clear zones formed were DU 4.2 (Candida sorboxylosa) and DU4.22 (Cyberlindnera fabianii) isolates with amylolytic index of DU 4.2 isolates at 0.24 and DU 4.22 at 0.72 with an incubation time of 48 h. The highest amylase enzyme activity was found in isolate DU 4.2 of 31.21 U / mL.
Yeast diversity, Durio kutejensis, Metagenomic, Amylase production, Solid State Fermentation (SSF)
Durian is a tropical fruit native to Southeast Asia, especially Indonesia, regarded as the “King of Fruits” due to its distinctive (large, covered by spines) shape and rich flavor. Scientifically, durian belongs to the group of Malvaceae, family Bombacaceae and genus Durio. Kostermans1 and David2 have recorded 27 species of durian, with the total of 19 species are found in Kalimantan, Indonesia and 11species in Malacca Peninsula, 7 species in Sumatra and 1 species in Myanmar. Among all, only 7 species are edible for human, which are Durio zibethinus (durian), Durio kutejensis (lai), Durio oxleyanus (kerantongan), Durio dulcis (lahong), Durio graveolens (labelak), Durio grandiflorus (durian monkey), and Durio testinarium (turtle durian)3,4. Durio zibethinus is the most widely cultivated by Indonesians because of its delicious taste and smooth texture. One of Indonesia’s original fruits from East Kalimantan is Lai Durian (Durio kutejensis)5 which has drier pulp, yellow-orange with distinctive aroma (not like durian) and less pungent6. However, information about Lai Durian is still limited.
Lai cultivars were reported to have less diverse sulfurs and esters as compared to the other durian, which were most probably the key reason for the different aroma characteristics8. Because of its distinctive flavor and aroma, lai becomes one of Indonesia’s tropical fruit commodities which is potentially developed as raw materials for fermented food and beverages, and have large domestic market opportunities and exports, especially in pharmaceutical industry10,. Besides aromatic compounds (Methyl acetate, Ethyl acetate, Methyl propionate, Ethyl propionate, n-Propyl propionate, Ethyl iso-butyrate, Ethyl butyrate, Methyl-methylbutyrate), ester compounds (Ethyl-2-methylbutanoate, Ethyl iso-butyrate, Ethyl butyrate, Methyl-methylbutyrate), ether compounds (Ethyl-2-methylbutanoate, volatile Ethyl acetate), and Sulphur compounds (Hydrogen sulphide, Methanethiol, Ethanethiol, Propanethiol)7. Lai consists of biomass containing hemicelluloses, and lignin which can be used as carbohydrate resources5.
Lai Durian (Durio kutejensis) with its distinctive flavor and aroma becomes one of Indonesia’s tropical fruit commodities which is potentially to be developed as raw materials for fermented food and drinks and have large domestic market opportunities and exports8. The main content of durian seeds is starch and protein9. Some researches on starch contained in durian seed have been carried out, including characterization10, hydrolysis with acids, and its application to as raw materials of nuggets and even bioplastic material. Like all durian seeds, Lai seeds also have the same source of starch, but the extraction of starch from its flesh and the measurement of functional properties have not been explored further.
The presence of chemical compounds in lai have been reported to be associated with endophytic microorganisms exist in the fruit. The physicochemical, microbial diversity and sensory profile of lai changes during post-harvest because the natural fermentation occurs. During fermentation, yeasts are responsible for material degradation from complex into simpler compounds. Yeasts like Saccharomyces spp. have been successfully isolated from durian fruit (Durio zhibetinus) and was used as starter inoculum for bioethanol production from cassava (Manihot esculentao18. Yeasts are single-celled microorganisms that can be found in various kind of fruits, such as Aureubasidium pullulans in grapes12; found in jackfruit13 with amylase enzyme activity (0.88 U/ mL); Pichia sp. from apple (Malus domestica) was also found14. Yeasts has been found to produce various enzymes for many industrial applications15-17. Yeasts like Saccharomyces spp. have been successfully isolated from durian fruit (Durio zhibetinus) and used as starter inoculum for bioethanol production from cassava (Manihot esculenta)18.
Metagenomics using next-generation sequencing is a technology that enables massive and parallel DNA sequencing. Called high throughput-sequencing, which means that in one time, this technology can run millions of DNA sequences from one sample of the same environment in a relatively short time. By using metagenomics, data can be gathered to develop a profile of yeast diversity and taxonomy19 In this study, we use metagenomics-based approach to obtain information of yeast diversity profile in post-harvested lai which might be associated with the metabolite secondary production, in this case is amylase enzyme20. Amylase enzymes can be obtained during fermentation done by yeast21,22 α-amylase enzyme is one of the enzymes used in the enzymatic hydrolysis process to convert starch to glucose. This enzyme cuts the α-1,4-glucoside bond specifically at a certain point to form dextrin11.
In this study, we also support the data with the rational reason for production optimization of amylase enzyme in Solid State Fermentation (SSF) by the culture-dependent24. This method has been widely used for optimal cultivation in low water activity condition24,25. We expect that this study will provide information about yeast diversity of lai and recommendation for alternative source of amylase enzyme for industrial needs. We hope that the result will be reliable and reproducible as guidance for experiments further.
Yeast isolation and total genomic extraction of Lai Durian (Durio kutejensis). A fresh-harvested lai fruit was collected from Desa Bukit Sawit, Central Kalimantan, Indonesia, and lai with specific criteria of good ripening, no deformed husks and normal odor and texture of pulp, were used in this study. Lai with the code of DU4 are chosen to be performed for yeast diversity analysis of whole pulp content using metagenomics approach. 0.5 g of Durian pulp was collected aseptically and was put into a conical tube. Total genomic DNA from samples were extracted using CTAB/SDS method. DNA concentration and purity was monitored on 1% agarose gel. The extracted DNA was then diluted to reach the concentration of 1 ng/μL using sterile water. The rRNA/ITS genes of distinct regions. All PCR reactions were carried out with Phusion® High-Fidelity PCR Master Mix (New England Biolabs). The PCR Products confirmation was done by mixing it with 1×loading buffer (contained SYB green) and was run for electrophoresis on 2% agarose gel for detection. The PCR products was followed by purification with Gene JETTM Gel Extraction Kit (Thermo Scientific). The reads were compared with the reference database (Silva database, https://www.arb-silva.de/) using UCHIME algorithm (UCHIME Algorithm, http://www.drive5.com/usearch/manual/uchime_algo.html) to remove the chimera sequences [The clean reads were finally obtained and the species annotation for each representative sequence was compared to the Silva Database (https://www.arb-silva.de/) based on Mothur algorithm to annotate taxonomic information. Phylogenetic relationship construction to obtain yeast diversity of different OTUs and the pre-dominant species in the sample was conducted using multiple sequence alignment by the MUSCLE software (Version 3.8.31 http://www.drive5.com/muscle/).
A fresh Lai fruit was collected from Desa Bukit Sawit, Central Kalimantan, Indonesia, and Lai durian with the code DU4 are chosen to perform yeast diversity analysis of whole pulp content using metagenomics approach. Durian with specific criteria such as good ripening, no deformed husks and normal odor and texture of pulp were used in this study. 0.5 g of Durian pulp was collected aseptically and was put into a conical tube. Total genomic DNA from samples were extracted using CTAB/SDS method. DNA concentration and purity was monitored on 1% agarose gel. According to the concentration, DNA was diluted to 1 ng/μL using sterile water. Amplicon Generation 16S rRNA/18S rRNA/ITS genes of distinct regions (16S V4/16S V3/16S V3-V4/16S V4-V5, 18S V4/18S V9, ITS1/ITS2, Arc V4) were amplified used specific primer (e.g. 16S V4: 515F-806R, 18S V4: 528F-706R, 18S V9: 1380F-1510R, et. al) with the barcode. All PCR reactions were carried out with Phusion® High-Fidelity PCR Master Mix (New England Biolabs). PCR Products Mixing and Purification Mix same volume of 1×loading buffer (contained SYB green) with PCR products and operate electrophoresis on 2% agarose gel for detection. PCR products was mixed in equidensity ratios. Then, mixture PCR products was purified with GeneJETTM Gel Extraction Kit (Thermo Scientific). The reads were compared with the reference database (Silva database, https://www.arb-silva.de/) [26] [2] using UCHIME algorithm (UCHIME Algorithm, http://www.drive5.com/usearch/manual/uchime_algo.html)27,3 to detect chimera sequences, and then the chimera sequences were removed28,4. Then the Clean Reads finally obtained. Species annotation for each representative sequence, the Silva Database (https://www.arb-silva.de/)26,2 was used based on Mothur algorithm to annotate taxonomic information. Phylogenetic relationship Construction In order to study phylogenetic relationship of different OTUs, and the difference of the dominant species in different samples (groups), multiple sequence alignment were conducted using the MUSCLE software (Version 3.8.31 http://www.drive5.com/muscle/).
Yeast isolation from Durio kutejensis using culture-dependent analysis
Lai with the code DU4 were collected to perform yeast isolation using culture-dependent analysis. Yeast isolation was done by inserting pieces of durian flesh into Erlenmeyer flask containing 45 mL of MEB medium and homogenized for 24 h. 0.1 mL of durian suspension and was diluted by 9.9 mL of sterile distilled water (10-2 dilution), followed by serial dilution until 10-6. 0.1 mL of 10-6 dilution was put into petri dish containing YMA pH 4 by spread technique. The pH was adjusted by adding 0.5 mL of 35% HCL to 300 ml of Yeast Malt Agar (YMA) media. The sample solution was flattened using the Drygalski spatula and incubated for 24 h at room temperature. The growing colonies were then purified to obtain pure culture on the YMA pH 4 medium with the quadrant streak method, which was incubated for 24-48h at room temperature; and carried out 2-3 times transfer until a single colony was obtained. The growing colonies of the yeast isolates were observed macroscopically after 48h as described by morphological characters such as color, texture, profile and margin were noted refereed to Sukmawati et al.29.
Screening of Potential Amylase-Producing Yeast Isolates
Qualitative assay for amylolytic activity was done by diffusion method [30. A total of 28 UNJCC yeast isolates from Lai durian fruit were spread by streak method to the YMA (Yeast Malt Agar) medium and was followed by incubation at 30°C for 72 h. Each isolate was then inoculated into YPSA (Yeast Peptone Starch Agar) medium with 1% starch addition and making wells. The wells were made using sterile straws on YPSA media which has been divided into 8 quadrants with 2 replications in each quadrant. One pick of each yeast isolates was collected and homogenized with 0.9 mL of sterile distilled water in Eppendorf tube. About 20μL of the yeast suspension was inoculated into YPSA media and was incubated at 30°C for 72 h. Amylase activity can be seen by the presence of clear zone around the colony on YPSA media after the addition of Lugol drops. The clear zone was then measured using analytic calipers. Amylolytic index was calculated31.
Inoculum and Medium Preparation for Solid State Fermentation (SSF)
The positive yeast isolates (DU4.2 and DU4.22) were inoculated into YMA slant agar medium by 15 streak methods, followed by incubation at room temperature for 48 h. The two isolates were then picked and put into 5 mL of distilled water, followed by homogenization using vortex. A total of 10% (v/w) of yeast suspension (0.5 mL in 5 grams) was put into a sterile fermentation medium. Fermentation medium composition was made in Erlenmeyer with 70% humidity as follows: 5 gram of wheat husk; 3.5 mL of nutrients (1% starch, 5 g peptone, 2 g yeast extract, 0.1 g CaCl2, 0.5 g KH2PO4 and 200 mg chloramphenicol) were dissolved in 1000 ml Mc’Ilvaine buffer (pH 3 – 5)24. The media is then homogenized using rod stirrer.
Optimization of Amylase Enzyme Extraction
Amylase enzyme extraction was carried out by solid state fermentation (SSF). Fermentation was run in incubator for 24h, 48h, 72h, and 96h at room temperature (28°C). Production of crude amylase enzyme extracts was carried out by adding 50 ml of sterile distilled water and agitation at 150 rpm rotator shaker for an hour (Oliveira et al. 2015). The medium was filtered using muslin cloth and Whattman paper and the crude enzyme extract were then put into the Falcon tube. The extract was centrifuged for 10 minutes at 3000xg and 4°C. The results obtained are in the form of filtrate (crude enzyme extract) and its sediment. The filtrate was expected to be a-amylase crude extract and collected for enzyme activity test based on Yalcin and Corbaci (2014) method. 0.5 ml of enzyme crude extract were placed in test tube and added with 0.5 ml of 1% starch solution containing Mcllavaine pH 4 buffer. The reaction was run in water bath with temperature of 60°C for 10 minutes. 1 ml of 3,5-dinitrosalicylate (DNS) reagent was then added to the tube. The enzymatic reaction was stopped by heating at 100°C for 5 minutes. The addition of 8 ml of sterile distilled water was carried out into the sample using pipette
Quantitative Amylolytic Activity Assay of Yeast Isolates
Quantitative amylolytic activity assay was performed based on absorbance using OD (Optical Density) spectrophotometer at wavelength of 540 nm. Amylase enzyme activity of each sample was tested three times based on33 method. Calculation of enzyme activity uses the formula34:
Molecular Identification of Isolate DU 4.2 and DU 4.22
Among 28 yeast isolates, isolate DU4.2 and isolate DU4.22 showed amylolytic activity and therefore were collected for molecular identification. The DNA yeast was extracted using the Genetic Plant Gneaid kit. DNA was amplified based on D1/D2 rDNA region using the following primers: forward- ITS5 (5’-GGAAGTAAAAGTCGTAACAAGG-3’) and reverse-ITS4 (5’TCCTCCGCTTATTGATATGC-3’). PCR cycling conditions for yeasts consisted of an initial denaturation step at 95°C for 2 min; 33 cycles of denaturation at 95°C for 15 sec, annealing at 58°C for 30 sec and extension at 68°C for 2 min; and a final extension at 68◦C for 10 min. The PCR product was purified with first base service. The sequences were aligned and compared with the NCBI database by the Internet using Basic Local Alignment Search Tool. Phylogenetic trees were made using MEGA 5 software with the Neighbor Joining (NJ) method. Phylogenetic trees are used to determine kinship relationships between species sampled with various other species. Molecular phylogenetic combines molecular biology techniques with statistics to reconstruct phylogenetic relationships.
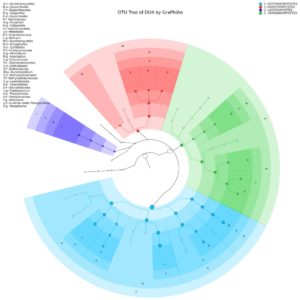
Yeast Diversity Information of Lai Durian Using Metagenomic Approach In this study, yeast diversity information was obtained based on ITS4 specific gene which is known to be conserve region of fungal groups. Next-Generation Sequencing (NGS) technology makes it possible to sequence millions of DNA fragments in one analysis at a relatively low cost compared to Sanger’s sequencing technology. Metagenome analysis was performed to see the diversity of culturable and unculturable microorganisms35. The sample of Lai durian with the code of DU4 was used for metagenomics analysis. This sample was chosen that is in good condition, without any damage and disease seen. The result of showed that only group of Ascomycota is present in the fruit with two genera consisting of four clade classes based on OTU analysis, which are Dothideomycetes, Eurotiomycetes, Leotiomycetes, and Sordariomycetes (Fig. 1). The class of Sordariomycetes dominated the yeast community, consisting of Botryosphaeria, Lasiodiplodia, Aureobasidium, Paraphoma, Preussia, Sporormiella. Lim T.K has reported some lesser known mycoflora of durian in Malaysia which are black mildew Ascomycetes species, Meliola durionis, and 7 sooty mould species: 3 from the Deuteromycetes and 4 from the Ascomycetes. Polychaeton sp., Leptoxyphium sp. and Tripospermum sp. belong to the former class while Scorias spongiosa, Phragmocapnias betle, Trichomerium grandisporum, and Trichopeltheca asiatica to the latter36.
Fig. 1. Yeast diversity information of Lai durian (Durio kutejensis) collected from Desa Bukit Sawit, Central Kalimantan, Indonesia, using metagenomic approach based on ITS4 gene. Only Ascomycota was present and there are four large phylum exist with the domination of Sordariomycetes (blue).
The dominant yeast present in durian fruit and its ability to produce volatile sulfur compounds (VSCs) has been found37. Durian is rich in carbohydrates, proteins, and fats relative to other fruits, allowing yeasts and molds to grow and develop. Botryosphaeria dothidea is a pathogenic yeast found in postharvest durian which is also found in fruits such as olives38. These yeast also produce volatile compounds such as fatty acids, esters, sterols and fatty acid methyl esters were some types of compounds recovered. Furthermore, the antioxidant capacity was measured, with the most promising result as 38.4±3.1 μmol TEAC/μg extract39. Lasiodiplodia crassispora, the species belongs to the family group of Botryosphaeriaceae, was also found, which can cause diseases in fruits such as fruit rot, leaf spots, dieback, cankers and root rot of Angiosperms and Gymnosperms worldwide40.
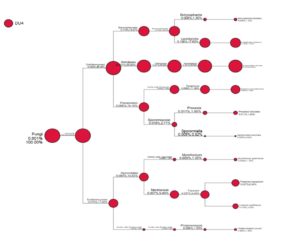
Fig. 2. Taxa diversity information of Lai durian (Durio kutejensis) collected from Desa Bukit Sawit, Central Kalimantan, Indonesia, using metagenomic approach based on ITS4 gene. 10 yeast species are present in the community.
As shown in Fig. 2, 10 species belong to the group of Ascomycota were identified as Botryosphaeria dothidea (0.008%,1.35%), Lasiodiplodia crassispora (0.106%,17.62%); Aureobasidium pullulans (0.331%,55.02%), Paraphoma chrysanthemicola (0.068%,11.38%), Preussia funiculate (0.011%,1.90%), Sporormiella intermedia (0.005%,0.82%), Myrothecium gramineum (0.008%,1.35%), Fusarium oxysporum (0.037%,6.24%), Fusarium proliferatum (0.019%,3.25%), and Phialemoniopsis curvata (0.006%,1.08%). From the result obtained, it can be seen that Aureobasidium pullulans accounted for most Ascomycota phylum present in Lai durian. Aureobasidium pullulans is a ubiquitous black, yeast-like fungus that can be found in different environments (e.g. soil, water, air and limestone). The discovery of some yeasts and molds in this study provides new information regarding the role and function of these microorganisms on durian fruit. The dominance of yeast A. pullulans benefits both fruit and human health. Aureobasidium pullulans plays an important role in the process of the food industry and the biocontrol of pathogenic molds in post-harvest fruits. Yeast like fungi A. pullulans has ability to produce amylase enzymes41; producing a-amylase42 acting as biocontrol agent controlling Fusarium sp.43; found in grapes44. The biocontrol activity of Aureobasidium pullulans has been reported in some researches; A. pullulans multiplied rapidly and controlled decay caused by either B. cinerea or P. expansum, reducing the incidence of gray and blue mold of apple by 89 and 67%, respectively, compared to the water-treated control45. Aureobasidium pullulans strain Ach1-1 was also successfully isolated for its biocontrol effectiveness against Penicillium expansum, the causal agent of blue mold on harvested apples46. Yeast species such as Saccharomyces cerevisae, Saccharomyces blourdeous, Zygosaccharomyces fermentatii, Candida sorboxylosa, Zygosaccharomyce bisporus, Saccharomyces blourdeous, can be found in many local fruit in Indonesia47.
Table (1):
Yeasts isolated from Lai durian DU4 on YMA medium, incubated at 28°C for 48h. 40 isolates were obtained with various morphological characteristics.
| Sample code | Number of isolates | Colony color | Colony texture | |||
|---|---|---|---|---|---|---|
| White | Smooth | Rough | Butyrous | Mucoid | ||
| DU4 | 28 | 28 | 26 | 2 | 1 | 27 |
| Total | 28 | 28 (100%) | 26 (92,85%) | 2 (7,14%) | 1 (3,57%) | 27 (96,42%) |
Culture-dependent Analysis of Yeasts Isolated from Lai Durian
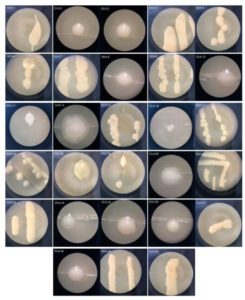
The results showed that from 3 collected Lai durian, 28 yeast isolates were successfully isolated (with code DU4) (Table 1). Based on morphological observation, all yeast isolated from Lai durian have white colony with mucoid and butyrous texture. The mucoid texture of the yeast was due to the presence of polysaccharide substances contained in the extracellular component of yeast48. Some yeast is covered by extracellular components in the form of slimy polysaccharides and heteropolysaccharides49. Research by Nyanga et al. reported that yeasts obtained from Masau (Ziziphus mauritiana) ripe have white to creamy colonies50. Research by Lentz et al. obtained all yeast isolates from Pindo palm fruit (Butia capitata), loquat (E. japonica), blackberry (Rubus sp.), and hackberry (Celtis sp.) have white to cream colonies51. Yeast can produce various kinds of pigments such as carotenoids and melanin but white yeast isolates indicate that yeast does not produce pigments. Carotenoid pigments are indicated in red to orange while black indicates melanin pigment52. Research Sukmawati et al. reported that yeast isolates obtained from the surface of Saeh leaves (Broussonetia papyrifera) were dominated by pigmented yeasts29. This might be caused by the leaf surface which experienced environmental stress. Leaf surfaces can be directly exposed to UV rays, air, and rain under varying conditions53.
Yeast can be found from a variety of substrates and has different roles either in amylase-producing or cellulose-producing isolates54,30,15,17, as well as controlling pathogenic fungi55. Research by Wulandari et al. has successfully isolated yeast from jackfruit and obtained 191 yeast isolates13. Amorim et al. obtained 132 yeast isolates from pineapple56. The population and yeast community on fruit is influenced by several factors such as: fruit type, geographical location, and fruit growth phase57. Yeast is rarely found in the fruit development phase and in immature fruit, but the number of yeast will increase when the fruit is ripe58. This is because ripe fruit contains more simple carbohydrates compared to unripe fruit, so the presence of yeast in ripe fruit is much more. The yeast isolates obtained were then purified to obtain single colony and were observed macroscopically (Fig. 3).
Fig. 3. Morphological characteristic of yeast isolated from Lai durian with code DU4, incubated on YMA medium at 28°C for 24 h.
Screening Results of Yeast Isolates Producing Amylase Enzyme
Screening results showed that there are 2 isolates (DU4.2 and DU4.22) showing the presence of clear zones around the well after Lugol drops, indicating that the two isolates are having amylolytic activity (Fig. 4). Furthermore, the isolates with positive amylolytic activity were tested using digital calipers and the amylolytic index values were calculated59. The amylolytic index of isolate DU4.2 and isolate DU4.22 are 0,24±0,57 and 0,72±0,39, respectively.
Fig. 4. The presence of clear zone appeared after Lugol drops by isolates DU4.2 (a) and isolates DU4.22 (b) incubated on YPSA medium at 30°C for 72 h.
The presence of amylase enzymes is indicated by the presence of a clear zone around the yeast isolate colony. Clear zone formed around the yeast colony shows that the isolate is able to hydrolyze starch. Starch will form a deep blue complex with iodine reagents. The iodine-starch reaction is caused by the presence of the helical amylose and iodine in forming I3– which place the helical core. Active hydrolysis of starch by amylase enzyme will cause the starch-iodine complex to decompose to form a clear zone. The absence of a clear zone around the colony indicates a reaction between the iodine reagent and the starch that is not hydrolyzed in the YPSA medium60. The ability or power to produce amylase enzymes in a microbe is characterized by the formation of clear zones in a medium containing starch61.
Morphological and Molecular Identification Result of Isolate DU4.2 and Isolate DU4.22
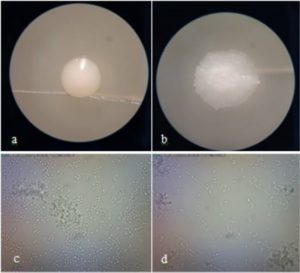
Identification of potential yeast probiotics was carried out by macroscopic and microscopic observations of isolate DU4.2 and isolate DU4.22 at 48 h. Macroscopic observations showed that both isolate DU4.2 and isolate DU4.22 showed white colony, mucoid texture, smooth colony surfaces, mounting colony profiles, and without hyphae (Fig. 5). Isolate DU4.2 has flat colony edge, cylindrical cell shape, and multilateral germination, while isolate DU4.22 has undulate colony edge, circle cell shape and germination at every colony edges. Phylogenetic trees are constructed to see the kinship of yeast species that have potential as probiotic agents. The results of the mapping of D1/D2 rDNA yeast isolates were aligned using the MUSCLE (Multiple Sequence Comparison by Log-Expectation) method62. The phylogenetic tree was constructed using the Neighbor Joining method with a bootstrap value of 1,000 replications63. Molecular identification of isolate DU4.2 and isolate DU4.22 was performed based on ITS DNA region. Results showed that isolate DU4.2 has the closest homology to Cyberlindnera fabianii NRRL 1871 with 100% homology, while isolate DU4.22 showed sequence homology of 99.16% to Candida sorboxylosa ADR2-2 (Table 2).
Table (2):
BLAST results of isolate DU4.2 and isolate DU4.22 based on D1/D2 rDNA region.
Isolate code |
NCBI result |
Max Score |
Query Cover |
E-value |
Accession |
Identity |
Gaps (%) |
|---|---|---|---|---|---|---|---|
DU4.2 |
Cyberlindnera fabianii NRRL 1871 |
1113 |
96% |
0,0 |
NG_055731.1 |
100% |
0/579
(0%) |
DU4.22 |
Candida sorboxylosa ADR2-2 |
675 |
93% |
0,0 |
MG833307.1 |
99,16% |
0/357
(0%) |
Fig. 5. Macroscopic observation of isolate DU4.2 (a) and isolate DU4.22 (b), and microscopic observation of isolate DU4.2 (c) and isolate DU4.22 (d)
Cyberlindnera fabianii is an ascomycetous ‘uncommon’ yeast which was previously named Candida fabianii64. The yeast of C. fabianii has a morphology in the form of a colony of creamy white cells, smooth surface, flat edges, round to oval shaped cells, and has budding65. This is consistent with macroscopic and microscopic observations of C. fabianii isolates which have macroscopic and microscopic characteristics similar to those of the yeast C. fabianii. Macroscopic observations showed that white C. sorboxylosa yeast isolate, mucoid textured, smooth colony surface, undulate colony edge and mountainous colony profile (Table 3). Microscopic observations showed that the isolate of yeast C. sorboxylosa has a round cell shape and buds at every place on the cell surface. The yeast of C. sorboxylosa has a morphology in the form of white cell colony, smooth surface, raised profile, and medium size66. This is consistent with macroscopic and microscopic observations of C. sorboxylosa isolates which have characteristics such as C. sorboxylosa.
Table (3):
Average absorbance value of of Cyberlindnera fabianii NRRL 1871 and Candida sorboxylosa ADR2-2 at 540 nm wavelength.
| Isolate code | Repetition | Incubation time (h) | |||
|---|---|---|---|---|---|
| 48 | 72 | 96 | 120 | ||
| Cyberlindnera fabianii NRRL 1871 | 1 | 1.05 | 1.04 | 1.26 | 0.96 |
| 2 | 1.18 | 1.08 | 0.87 | 0.93 | |
| 3 | 1.25 | 1.02 | 1.09 | 0.84 | |
| 4 | 1.15 | 1.19 | 0.92 | 0.54 | |
| Cyberlindnera fabianii NRRL 1871 | 1 | 0.79 | 0.63 | 0.56 | 0.89 |
| 2 | 0.53 | 0.55 | 0.58 | 0.77 | |
| 3 | 0.73 | 0.66 | 0.45 | 0.82 | |
| 4 | 0.58 | 0.66 | 0.45 | 0.49 | |
The phylogenetic tree showed that the isolate DU4.2 was in monophyletic clade with Cyberlindnera fabianii NRRL 1871 with bootstrap value of 82. Isolate DU4.22 was in a monophyletic clade with Candida sorboxylosa ADR2-2 with bootstrap value of 100 (Fig. 5). Bootstrap value in phylogenic trees shows the tree topology formed. Bootstrap value of 70-100% is a value that has a high level of confidence in the phylogenic tree produced67.
Cyberlindnera fabianii NRRL 1871, reviously Hansenula fabianii, Pichia fabianii, and Lindnera fabianii), has been proposed to be novel gene after multigene sequence analysis68. Candida sorboxylosa ADR2-2 has been diversely colonized palm wines, among which some were related to a specific type of wine and the majority of them have the ability to digest starch, sugar, protein or lipid69. PCR-restriction fragment length polymorphism of ITS-5.8S rDNA combined to 26S rRNA gene and/or the partial ACT1 gene sequencing were applied for yeast characterization, and their enzymatic profiles assessed by using API ZYM kits. Thirteen genera and 23 species were identified, with the highest diversity (14 species) in raffia wine. Saccharomyces cerevisiae was dominant and common to all palm wines. Some potentially pathogenic yeasts were also isolated. The majority of tested strains displayed high amylo-peptidase, phosphatase, β-glucosidase and a-glucosidase activities and esterase activity.
Fig. 6. Phylogenetic tree of isolate DU4.2 and isolate DU4.22, constructed using MEGA 5 software with the Joining Neighbor (NJ) method (1000x Bootstrap). Aspergillus niger was used as outgroup of the tree and the sequence collection were taken from NCBI.
Amylase Enzyme Activity of Cyberlindnera fabianii NRRL 1871 and Candida sorboxylosa ADR2-2 in Solid State Fermentation (SSF)
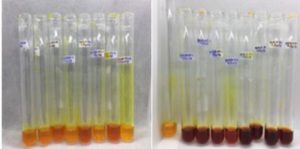
The fermentation medium with the SSF method is used in this study because it is one of the factors that influence the activity of amylase enzyme. Research conducted by Bhatti et al. obtained the optimum humidity to produce amylase enzyme at 70% humidity70. High humidity in the medium will cause the gas phase to be reduced and gas exchange to be hampered, causing the substrate to be anaerobic, while too low humidity will lead to poor microbial growth and decreased levels of nutrient acquisition71. After DNS reagent (3,5-dinitrosalicylate) application, the results of enzyme extract in solid state fermentation turned red as shown in Fig. 7. The principle of the DNS method with starch substrate is to reduce 3.5 dinitrosalicylate to 3-amino-5-nitrosalicylate resulting in a change from yellow to brownish red32. These reagents are commonly used to measure reducing sugars produced by microbes because of their high level of accuracy so that they can be applied to even small amounts of sugar. In an alkaline atmosphere reducing sugars will reduce 3,5-dinitrosalicylic acid (DNS) to form compounds that can be measured for absorption by a spectrophotometer at a wavelength of 540-550 nm72.
Fig. 7. Filtrate with the addition of 1% starch extract and DNS reagents before heating (left) and filtrate after heating (right).
The results of testing the activity of the amylase enzyme required calibration to determine glucose levels by making a glucose standard curve. Glucose solution was chosen as the solution for making standard curves because glucose includes reducing sugars produced from hydrolysis of the substrate by the amylase enzyme. The results of the standard curve analysis have a linear equation y = 0.3889x + 0.0619 with a correlation value (R2) of 0.9772. The equation obtained is used to determine the concentration of glucose in the test sample amylase enzyme activity. Correlation value (R2) which is close to number 1 shows the correlation between absorbance value and glucose concentration73. Table 3 showed the average absorbances of Cyberlindnera fabianii NRRL 1871 and Candida sorboxylosa ADR2-2 at wavelength 540 nm.
Table (4):
Amylase activity value of Cyberlindnera fabianii NRRL 1871 and Candida sorboxylosa ADR2-2 at pH 4 in Solid State Fermentation method.
| Isolate code | Incubation time (h) |
Amylase Activity (U/mL) (Mean±SE) |
|---|---|---|
| Cyberlindnera fabianii | 24 | 25.32b ± 1.84 |
| 48 | 31.21c ± 1.20 | |
| 72 | 29.81bc ± 1.11 | |
| 96 | 27.32bc ± 2.52 | |
| Candida sorboxylosa | 24 | 14.08 a ±1.45 |
| 48 | 17.06 a ±1.72 | |
| 72 | 16.07 a ±0.73 | |
| 96 | 12.73 a ±0.97 |
Table 4 showed that based on the measurement of amylase enzyme activity, Cyberlindnera fabianii NRRL 1871 has higher value (31.21 U/mL; 48 h incubation time) than Candida sorboxylosa ADR2-2 (12.73 U/mL; 96 h incubation time). This result showed that there is an effect of the incubation time of solid state fermentation with pH 4 on the activity of the amylase enzyme produced by both isolates.
Both isolates produced the highest activity at 48 hours of incubation. Enzyme activity or enzyme work has U/ml units. One unit (U) of amylase enzyme activity is defined as the amount of enzyme that produces 1 µmol reducing sugar (glucose) per minute under conditions of enzyme activity testing (Lehninger, 2008). Martin et al. (1983) stated that enzyme activity is strongly influenced by the incubation time. The incubation time is the time required by the enzyme to interact with the substrate, if the enzyme is saturated with the substrate, the enzyme will not work optimally. Darwis (1995) also states that at the beginning of fermentation the enzyme activity is still very low. Enzyme activity will increase with increasing fermentation time. This follows the growth pattern of microorganisms that experience several growth phases, namely the adaptation phase, the exponential phase, the stationary phase, and the death phase. Amylase enzymes are produced by yeast in the adaptation phase and reach optimum during the final exponential phase (Sjofjan and Ardyati 2011). Amylase enzyme activity will decrease after the yeast reaches the final exponential phase because the starch in the fermentation medium starts to run out so the enzyme cannot be produced. Yeast plays a role in the process of maturation and control of pathogenic molds on durian fruit. Amylase activity increased at 48 hours incubation.
Research on the yeast Candida sorboxylosa and Cyberlindnera fabianii as amylase enzyme producers is still limited. Candida sorboxylosa isolated from local fruits of Ethiopia can ferment the type of fructose sugar present in bread dough47. Sugar breakdown that occurs will release carbon dioxide in the bread dough. This shows that Candida sorboxylosa has an enzyme that is responsible for fermenting most of the sugar in the bread dough. While the yeast Cyberlindnera fabianii has been known to have acetate ester hydrolase enzyme activity and can produce ethanol78,79.
Metagenomic analysis result showed that yeast isolated from Lai durian (Durio kutejensis) collected from Bukit Sawit Village in Central Kalimantan, Indonesia was associated with Ascomycota phylum, consisting of 10 species including Botryosphaeria dothidea (0.008%, 1.35%), Lasiodiplodia crassispora (0.106%, 17.62%); Aureobasidium pullulans (0.331%, 55.02%), Paraphoma chrysanthemicola (0.068%, 11.38%), Preussia funiculate (0.011%, 1.90%), Sporormiella intermedia (0.005%, 0.82%), Myrothecium gramineum (0.008%, 1.35%), Fusarium oxysporum (0.037%, 6.24%), Fusarium proliferatum (0.019%, 3.25%), and Phialemoniopsis curvata (0.006%, 1.08%). Culture-dependent analysis showed that among 40 yeasts isolated from 3 Lai durian samples, isolate with the code DU4.2 and DU4.22 exhibited the clear zones with amylolytic activity value at 0.24 and 0.72. Molecular identification resulted that these two isolates have the closest homology to Cyberlindnera fabianii NRRL 1871 and Candida sorboxylosa ADR2-2, respectively. Isolate DU4.2 Cyberlindnera fabianii NRRL 1871 has higher amylase activity value (31.21 U/mL) with an incubation time of 48h at pH 4 than Candida sorboxylosa ADR2-2 (12.73 U/mL) with 96 h incubation time. This research was a study describing yeast information that could be found from various native fruits of Indonesia, especially Lai durian. Yeast isolated from Lai durian are able to produce amylase enzyme which can be potentially developed in industrial application.
ACKNOWLEDGMENTS
The authors are very grateful to DRPM Kemenristekdikti, Hibah Penelitian Terapan Unggulan Perguruan Tinggi (PTUPT) 2021-2022 on behalf Dalia Sukmawati with title: “Aplikasi khamir probiotik untuk pengembangan prototipe sentra produksi kakao Indonesia berkualitas ekspor. We thank the Lab. Microbiology and Universitas Negeri Jakarta Culture Collection (UNJCC) for the facilities provided to run this study.
CONFLICT OF INTEREST
The authors declares that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
DS, HEE conceived and designed the experiments; SN and ZN performed the experiments; DS, RW, DJD, analyzed the data; DS, SNA, HEE, DJD wrote the paper.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files.
- Kosterman AJGH. Notes on Durian (Durio) Species of East Borneo[M]. Kalimantan: De Tropische Natur. 1953:31-35.
- Baum DA, Alverson WS, Nyffler R. A durian by any other name: taxonomy and nomenclature of the core Malvales. Cambridge: Harvard Papers in Botany. 1998,3(2):315-330.
- Aziz NAA Aziz, Jalil AMM. Bioactive Compounds, Nutritional Value, and Potential Health Benefits of Indigenous Durian (Durio zibethinus Murr.): A Review [J]. Foods. 2019;8(3):96.
Crossref - Wisutiamonkul A, Promdang S, Ketsa S, van Doorn WG. Carotenoids in Durian Fruit Pulp during Growth and Postharvest Ripening. Food Chemistry. 2015;180:301-305.
Crossref - Faisal M, Gani A, Maulana F. Preliminary Assessment of the Utilization of Durian Peel Liquid Smoke as a Natural Preservative for Mackerel. F1000Research. 2019;8:240.
Crossref - Sri S. Antarlina. Identification of Physical and Chemical Properties of Local Fruits in Kalimantan. Buletin Plasma Nutfah. 2009;15(2):80-90.
Crossref - Josilene Lima Serra, Fabio Gomes Moura, Gilberto V. de Melo Pereira, Soccol CR, Darnet S. Determination of the Microbial Community in Amazonian Cocoa Bean Fermentation by Illumina-Based Metagenomic Sequencing. Food Science and Technology. 2019;106: 229-239.
Crossref - Z.A. Mohamed, M.S. Othman, A.T. Sapii, Z. Mahmood, and S. Idris, Diversity Durio at Sarawak Prosiding Durian 2000: Quality and marketing 2000. Malaysia: 2000:212-215.
- Contarino R, Brighina S, Fallico B, Cirvilleri G, Parafati L, Restuccia C. Volatile Organic Compounds (VOCs) Produced by Biocontrol Yeasts. Food Microbiology. 2019;82:70-74.
Crossref - Soebagio B, Sriwidodo, Adhika AS. Pengujian sifat fisikokimia pati biji durian (Durio zibethinus Murr) alami dan modifikasi secara hidrolisa asam. Bandung: Universitas Padjajaran, 2009.
- Arung ET, Suwinarti W, Hendra M, et al. Determination of Antioxidant and Anti-Melanogenesis Activities of Indonesian Lai, Durio kutejensis [Bombacaceae (Hassk) Becc] Fruit Extract. Tropical Journal of Pharmaceutical Research. 2015;14(1):41-46.
Crossref - Lorenzini M, Zapparoli G. Yeast-like Fungi and Yeasts in Withered Grape Carposphere: Characterization of Aureobasidium pullulans Population and Species Diversity. Int J Food Microbiol. 2019;289:223-230.
Crossref - Tria Putri Wulandari, Dalia Sukmawati, Tri Handayani Kurniati. Isolasi Dan Seleksi Khamir Amilolitik Asal Buah Nangka (Artocarpus heterophyllus Lam.). Bioma. 2018;13(1):37-42.
Crossref - Fadhli H, Kusdiyantini E, Nurhayati. Karakterisasi morfologi , biokimia, dan uji enzimatis isolat khamir buah apel (Malus domestica Borkh.) yang berpotensi menghasilkan bioetanol. Biologi Tropika. 2019;2(2):62-73.
- Risandi A, Sukmawati D, Jusna SNA, enshasy HE, Ridawai R. Isolation and Screening of Amylolytic Yeast from Paphiopedilum Sp., Originating from Bedugul Botanical Garden, Bali, Indonesia. Journal of Physics: Conference Series. 2019;1402:1-6.
Crossref - Sukmawati D, Dellanerra D, Risandi A. Screening the Capabilities of Indonesian Indigenous Mold in Producing Cellulase Enzyme. IOP Conference Series: Materials Science and Engineering. 2018;434(1).
Crossref - Sukmawati D, Larasati RP, Kurniati TH, et al. Molds Isolated from Chicken Feed as Potential Amylase Resources. International Journal of Scientific and Technology Research. 2019;8(11):188-196.
- Hermansyah, Tounaly Xayasene, Nguyen Huu Tho, Fatma, Panagan AT. Bioethanol Production from Cassava (Manihot esculenta) Peel Using Yeast Isolated from Durian (Durio zhibetinus). Journal of Physics. 2018;1095:012016.
Crossref - Robibins RJ, Krishtalka L, Wooley JC. Advances in biodiversity: metagenomics and the unveiling of biological dark matter. Standards in Genomic Sciences. 2016;11(1):69.
Crossref - Teeling H, Glockner FO. Current opportunities and challenges in microbial metagenome analysis-a bioinformatic perspective. Briefings in Bioinformatics. 2012;13(6):728-742.
Crossref - Pangastuti A, Wahjuningrum D, Suwanto A. Isolasi, Karakterisasi, dan Kloning Gen Penyandi α-Amilase Bakteri Halofil Moderat Asal Bledug Kuwu Isolation, Characterization, and Cloning of the α -Amylase Gene. Hayati. 2002;9(1):10-14.
- Yu W, Zou W, Dhital S, et al. The Adsorption of a-Amylase on Barley Proteins Affects the in Vitro Digestion of Starch in Barley Flour[J]. Food Chemistry. 2018;241:493-501.
Crossref - Hargono. Pemanfaatan Umbi Gadung Beracun (Dioscorea hispida) Sebagai Bahan Baku Pembuatan Bioetanol Untuk Bahan Bakar Kompor Rumah Tangga: Perancangan Distilasi Satu Tahap [C]// Prosiding Seminar Nasional Teknik Kimia “Kejuangan”. Yogyakarta:2015:1-7.
- de Oliveira APA, Silvestre MA, Alves-Prado HF, et al. Bioprospecting of yeasts for amylase production in solid state fermentation and evaluation of the catalytic properties of enzymatic extracts. Afr J Biotechnol. 2015;14(14):1215-1223.
Crossref - Ravichandran S, Vimala R. Solid State and Submerged Fermentation for the Production of Bioactive Substances : A Comparative Study. International Journal of Sciene and Nature. 2012;3(3):480-86.
- Illeghems K, De Vuyst L, Papalexandratou Z, Weckx S. Phylogenetic analysis of a spontaneous cocoa bean fermentation metagenome reveals new insights into its bacterial and fungal community diversity. PLoS ONE, 2012;7(5):e38040.
Crossref - Nielsen DS, Teniola OD, Ban-Koffi L, Owusu M, Andersson TS, Holzapfel WH. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int J Food Microbiol. 2007;114(2):168-186.
Crossref - Lisdiyanti P, Katsura K, Potacharoen W, et al. Diversity of acetic acid bacteria in Indonesia, Thailand, and the Philippines. Microbiol Cult Coll. 2003;19:91e98.
- Sukmawati D, Oetari A, Hendrayanti D, et al. Identification of phylloplane yeasts from paper mulberry (Broussonetia papyrifera (L.) L’Her. ex Vent.) in Java, Indonesia. Malays J Microbiol. 2015;11(4):324-340.
Crossref - Sukmawati D, Arman Z, Sondana G, et al. Potential Amylase-Producing Yeast Isolated from Indigenous Fermented Beverages Originating from Bali, Indonesia. Journal of Physics: Conference Series. 2019;1402(5).
Crossref - Goldbeck R, Andrade CCP, Pereira GAG, Filho FM. Screening and Identification of Cellulase Producing Yeast-like Microorganisms from Brazilian Biomes. African Journal of Biotechnology. 2012;11(53):11595-11603.
Crossref - Yalcin HT, Corbac C. Isolation and Characterization of Amylase Producing Yeasts and Improvement of Amylase Production[J]. Turkish J Biochem. 2013;38(1):101-108.
Crossref - Ajayi AO, Fagade OE. Heat Activation and Stability of Amylases from Bacillus Species. African Journal of Biotechnology. 2007;6(10):1181-1184.
Crossref - Aktar MB, Islam MS, Rahman MM. Determination of alpha-amylase activity of Streptomyces spp isolated from Bangladeshi soils[J]. International Journal Interdiscipline Multidiscipline Study. 2014;1(10):167-170.
- Boumehira AZ, Arous O, Elsayed EA, et al. Application of Attenuated Total Reflection/Fourier Transform Infrared Spectroscopy in the Screening of Strains Producing Bioactive Molecules: A Metabolomics Approach. Journal of Scientific and Industrial Research. 2019;78(5):301-306.
- Kwee LT. Studies on some lesser known mycoflora of durian: sooty mould and black mildew. Pertanika.1989;12(2):159-166.
- lu Y, Fong ASYL, Jian Yong Chua, et al. The Possible Reduction Mechanism of Volatile Sulfur Compounds during Durian Wine Fermentation Verified in Modified Buffers. Molecules. 2018;23(6):1456.
Crossref - Latinovic J, Mazzaglia A, Latinovi N, Ivanovic M, Gleason ML. Resistance of Olive Cultivars to Botryosphaeria Dothidea, Causal Agent of Olive Fruit Rot in Montenegro. Crop Protection. 2013;48:35-40.
Crossref - Valente IdeL, Confortin TC, Luft L, et al. Extraction of Bioactive Compounds from Botryosphaeria Dothidea Using Supercritical Carbon Dioxide and Compressed Liquefied Petroleum Gas. Journal of Supercritical Fluids. 2018;136:52-59.
Crossref - Phillips AJL, Alves A, Abdollahzadeh J, et al. The Botryosphaeriaceae: Genera and Species Known from Culture. Studies in Mycology. 2013;76:51-167.
- Negi S, Vibha K. Amylolytic Enzymes Current Developments in Biotechnology and Bioengineering. Elsevier. 2017:25-46.
- Homaei A, Ghanbarzadeh M, Monsef F. Biochemical Features and Kinetic Properties of a-Amylases from Marine Organisms. Int J Biol Macromol. 2016;83:306-314.
Crossref - Sarrocco S, Vannacci G. Preharvest Application of Beneficial Fungi as a Strategy to Prevent Postharvest Mycotoxin Contamination: A Review. Crop Protection. 2018;110:160-170.
Crossref - Clavijo A, Isabel L. Calderon, Paneque P. Diversity of Saccharomyces and Non-Saccharomyces Yeasts in Three Red Grape Varieties Cultured in the Serrania de Ronda (Spain) Vine-Growing Region. Int J Food Microbiol. 2010;143(3):241-245.
Crossref - Ippolito A, El-Ghaouth A, Wilson CL, Wisniewski M. Control of postharvest decay of apple fruit by Aureobasidium pullulans and induction of defense responses. Postharvest Biol Technol. 2000;19(3):265-272.
Crossref - Bencheqroun SK, Bajji M, Sebastien M, et al. Biocontrol of blue mold on apple fruits by Aureobasidium pullulans (strain Ach 1-1): in vitro and in situ evidence for the possible involvement of competition for nutrients. Commun Agric Appl Biol Sci. 2006;71(3 Pt B):1151-1157.
- Tsegaye Z, Tefera G, Gizaw B, et al. Characterization of Yeast Species Isolated from Local Fruits Used for Bakery Industrial Application Article Information. Journal of Applied Microbiological Research. 2018;1(1):21-26.
- Marham HD, Rustam Y, Sukmawati D. Uji Kemampuan Antagonisme Khamir Asal Daun Jati (Tectona grandis) Terhadap Kapang Pengkontaminan Pada Pakan Ternak Ayam. Bioma. 2017;12(2):118-125.
Crossref - Fardiaz S. Mikrobiologi Pangan 1. Jakarta: PT Gramedia Pustaka Utama. 1992
- Nyanga LK, Nout MJR, Gadaga TH, Theelen B, Boekhout T, Zwietering. Yeasts and Lactic Acid Bacteria Microbiota from Masau (Ziziphus mauritiana) Fruits and Their Fermented Fruit Pulp in Zimbabwe. Int J Food Microbiol. 2007;120(1-2):159-166.
Crossref - Lentz M, Putzke T, Hessler R, Luman E. Genetic and Physiological Characterization of Yeast Isolated from Ripe Fruit and Analysis of Fermentation and Brewing Potential. Journal of the Institute of Brewing. 2014;120(4):559-564.
Crossref - Dufosse L. Current and Potential Natural Pigments From Microorganisms (Bacteria, Yeasts, Fungi, Microalgae). Handbook on Natural Pigments in Food and Beverages: Industrial Applications for Improving Food Color. Woodhead Publishing, 2016:337-354.
Crossref - Deak T. Environmental Factors Influencing Yeasts In Book Biodiversity and Ecophysiology of Yeasts. Berlin: Springer. 2006:155-174.
Crossref - Dellanerra D, Risandi A, Sunari A, et al. Screening and Characterization of Amylolitic Mold Originated from Ghost Crab (Ocypode Sp.) in Cidaon, Ujung Kulon National Park, Indonesia. AIP Conference Proceedings. Indonesia. 2019:1-9.
Crossref - Sukmawati D. Antagonism Mechanism of Fungal Contamination Animal Feed Using Phylloplane Yeasts Isolated from the Bintaro Plant (Cerbera manghas) Bekasi in Java, Indonesia. Int J Curr Microbiol Appl Sci. 2016;5(5):63-74.
Crossref - Amorim JC, Piccoli RH, Duarte WF. Probiotic Potential of Yeasts Isolated from Pineapple and Their Use in the Elaboration of Potentially Functional Fermented Beverages. Food Res Int. 2018;107:518-527.
Crossref - Fleet GH. Yeast interactions and wine flavour. Int J Food Microbiol. 2003;86(1-2):11-22.
Crossref - Vadkertiova R, Molnarova J, Vranova D, Slavikova E. Yeasts and yeast-like organisms associated with fruits and blossoms of different fruit trees. Can J Microbiol. 2012;58(12):1344-1352.
Crossref - Soeka YS. Optimization and Characterization of Amylase from Actinomycetes Isolates From East Kalimantan. Berita Biologi. 2010;10(3):361-67.
- Sukmawati D, Setyaningsih A, Rahayu S, et al. Isolation and characterization of aflatoxigenic Aspergillus spp. from maize of livestock feed from Bogor. In IOP Conference Series: Materials Science and Engineering. 2018;434(1):012105). IOP Publishing.
Crossref - Gana T, Noel H, Mendoza BC, Monsalud RG. Isolation, Screening and Characterization of Yeasts with Amyloytic, Lipolytic, and Proteolytic Activities from the Surface of Philippine Bananas (Musa spp.). Philippine Journal of Science. 2004;143(1):81-87.
- Edgar RC. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinformatics. 2004;5(113):1-19.
Crossref - Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
- Arastehfar A, Fang W, Al-Hatmi AMS, et al. Unequivocal Identification of an Underestimated Opportunistic Yeast Species, Cyberlindnera fabianii, and Its Close Relatives Using a Dual-Function PCR and Literature Review of Published Cases. Medical Mycology. 2019;57(7):833-840.
Crossref - Ratnaningtyasa NI, Hernayantia, Ekowatia N, Sukmawati D, Widianti H. Chicken drumstick mushroom (Coprinus comatus) ethanol extract exerts ahypoglycaemic effect in the Rattus norvegicus model of diabetes. Biocatalysis and Agricultural Biotechnology. 2019;19:101050.
Crossref - Tsegaye Z. Isolation, Identification and Characterization of Ethanol Tolerant Yeast Species from Fruits for Production of Bio-ethanol. International Journal of Modern Chemistry and Applied Science. 2016;3(3):437-443.
- Michael Simpson. Plant systematics [M]. Burlington : Elsevier. 2006.
- Kurtzman CP, Robnett CJ, Basehoar-Powers E. Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Research. 2008;8(6):939-954.
Crossref - Djeni TN, Kouame KH, Ake FDM, et al. Microbial diversity and metabolite profiles of palm wine produced from three different palm tree species in Cote d’Ivoire. J Appl Microbiol. 2020;126(2):567-579
- Bhatti H, Hamid M, Nawaz R, et al. Optimization of Media for Enhanced Glucoamylase Production in Solid-State Fermentation by Fusarium solani. Food Technology and Biotechnology. 2007;45(1): 51-56.
- Kaur H, Arora M, Bhatia S, Alam MS. Optimization of a-Amylase and Glucoamylase Production in Solid State Fermentation of Deoiled Rice Bran (DRB) by Rhizopus oryzae. International Journal of Pure & Applied Bioscience. 2015;3(6):249-256.
Crossref - Apriyantono A, Fardiaz D, Puspitasari NL, et al. Analisis Pangan. Bogor : IPB Press. 1989.
- Mustika S, Rahmad M, Qadir A. Kemunduran Benih Kedelai Akibat Pengusangan Cepat Menggunakan Alat IPB 77-1 MM Dan Penyimpanan Alami. Journal of Chemical Information and Modeling. 2019;53(9):1689-1699.
- Lehninger AL. Principle of Biochemistry Ed 5. New York (US): WH Freeman and Company. 2008.
- Kukor JJ, Martin MM. Acquisition of digestive enzymes by siricid woodwasps from their fungal symbiont. Science. 1983;220(4602):1161-1163.
Crossref - Darwis, Sailah, Safriani, et al. Kajian kondisi fermentasi pada produksi selulase dari limbah kelapa sawit (tandan kosong dan sabut) oleh Neurospora sitophila. J Teknologi Industri Pertanian. 1995;5(3):199-207.
- Sjofjan O, Ardyati T. Extracellular Amylase Activity of Amylolytic Bacteria Isolated from Quail’s {Coturnix japonica) Intestinal Tract in Corn Flour Medium. Worlds Poult Sci J. 2011;10(5):411-415.
Crossref - van Rijswijck I, Kruis AJ, Wolkers-Rooijackers JCM, Abee T, Smid EJ. Acetate-Ester Hydrolase Activity for Screening of the Variation in Acetate Ester Yield of Cyberlindnera fabianii, Pichia kudriavzevii and Saccharomyces cerevisiae. LWT-Food Science and Technology. 2019;104:8-15.
Crossref - van Rijswijck IMH, Wolkers-Rooijackers JCM, Abee T, Smid EJ. Performance of Non-Conventional Yeasts in Co-Culture with Brewers’ Yeast for Steering Ethanol and Aroma Production. Microbial Biotechnology. 2017;10(6):1591-1602.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.