ISSN: 0973-7510
E-ISSN: 2581-690X
Food and water contaminations with heavy metals have been increasing due to the environmental pollution. Decontamination of mercury as one of the most toxic heavy metals seems necessary. The aim of this study is to use L. acidophilus ATCC 4356 to reduce the mercury amount in milk. All possible process variables (including contact time, bacterial count, mercury concentration, temperature, contact time and shaking rate) were screening by Plackett Burman design for determination of main effects. Then main effects (contact time, as well as Hg and biomass concentration) were studied in 5 levels with response surface methodology to reach maximal bioremoval efficiency. The highest decontamination efficiency (72%) was achieved in the presence of 80 μg/L of initial Hg concentration, 1 × 1012 CFU of L. acidophilus ATCC 4356 in the 4th day. Finally, the capacity of this bacterium for Mercury bioremoval was determined at different Hg initial concentrations by using the isotherm models of Langmuir and Freundlich. The results showed the higher correlation coefficient in Langmuir model so, Mercury absorptions obey Langmuir isotherm model. This study indicated that in the case of milk contamination to Hg, as reported in some countries, one of the solutions for metal decontamination could be the bioremoval by lactobacillus as natural valuable biosorbents as an environmental friendly technology.
Lactobacillus acidophilus, Mercury, Biosorbent, Removal, Milk, Isotherm
Heavy metals with the density of more than 5 g/cm3 are the essential elements for human body such as Zinc, Iron and Copper whereas some others are toxic even in very low amounts (in the range of μg/L) like Lead, Cadmium, Arsenic and Mercury1. There is an unwanted increasing in pollution of heavy metals into the air, water and soil and therefore food2,3. The sources of Mercury pollution are classified as natural resources (weathering of rocks, volcanic activities and biological processes), and the industrial activities (electricity power stations, mining, production of chemical, pesticides, cement, chlorine, mirrors, medical equipment and wastewater4.
Milk is well-known all around the world for having vital effects on human health. The level of toxic metals is an important issue in quality and safety of milk5. Mercury is one of the toxic metals widely spread in the environment, water and food6. Mercury is recognized as human carcinogenic metal and produces gastrointestinal and immunological disorders7. The scientific food safety agencies are responsible for human health. Codex standard for contaminants and toxins in food has allowed the maximum permissible limits for mercury concentration in milk as less than 0.05 µg/L8. There are some reports of mercury contamination in milk in some countries like China 0.08 µg/L9 and Iran, 0.07 µg/L10.
Chemical and biological techniques have been used to eliminate heavy metals from polluted solutions. Chemical process like using rezin11, ion exchange12 and nanomaterials13 but they are not efficient for low concentrations of the heavy metals and also are expensive and not environmentally safe.
Biological methods include using biosorbents like plants and microorganisms e.g. yeasts, bacteria, algae and fungi for bioremoval of heavy metal from food and water14-16.
In the previous reports of this experimental team, we used Saccharomyces cerevisiae to remove heavy metals from water17 and milk18,19 but in this novel project the biosoption of mercury by L. acidophilus is evaluated as Lactic acid bacteria (LAB) are popular probiotics using all over the world.
The LAB have a desirable background of using in food processes in a safe manner as they are in the list of generally recognized as safe15. LABs have been reported to have the possibility in health applications and also bind the food contaminations like heavy metals and toxins even in low concentrations. Negative surface charge of LABs facilitates the binding to cations. So LAB would be a great microorganism for using in reduction heavy metals in water and foodstuffs20,21.
LABs have been reported to remove the heavy metals bioadsorption of Cr by a novel Bacillus sp. CRB-B122, Hg bioremoval by L. acidophilus6, biosorption of As by Bacillus ferrooxidans23, Cd bioremoval by Bacillus coagulans and L. plantarum24, Hg bioremoval by B. cereus25, Se uptake by L. acidophilus 26 and As removal by L. acidophilus27. The gap of research in the previous reports, is the lack of an experimental design to evaluate all process variables for removal of heavy metal in the foodstuff and water (µg/L levels) instead of removal of heavy metal in wastewater (mg/L levels).
The aim of this study was to evaluate the capability of L. acidophilus ATCC 4356 for removing of mercury in milk in the range of µg/L. So, at first all possible process variables (including contact time, bacterial concentration, mercury concentration, temperature, contact time and shaking rate) were listed for a screening method of Plackett Burman design (PBD) and determination of main effects. Then, the main variables (contact time, as well as Hg and biomass concentration) have been studied in in 5 levels in response surface methodology (RSM) to reach the optimum condition for bioremoval efficiency. Finally, the capacity of L. acidophilus for Mercury bioremoval was determined at different Hg initial concentrations and also the biosorption isotherms were evaluate by using the two most famous isotherm models: Langmuir and Freundlich.
To our knowledge, there is no published study about the capability of L. acidophilus in biosorption of mercury in milk and this would be the first step of applying this valuable microorganism to remove the low levels (µg/L) of Hg concentration in milk successfully, therefore these results would open a new window in food decontamination by using this green technology for heavy metals removal in food industry.
Reagents and chemicals
The standard solution of Hg (NO3)2 (1000 mg/L, Merck, Spain) and MRS agar, MRS broth and plate count agar were obtained from Liofilchem (Zona Industriale, Italy). The other chemicals were: nitric acid (Merck), phosphate-buffered saline (HyClone, Spain), H2O2 (Prolabo, Spain) and bovine serum albumin (Labclinic, Spain).
Bacterial strain and preparation
L. acidophilus ATCC 4356 as one of the most available and widely used probiotic was selected and prepared from Tak Gen Zist Company (Tehran, Iran). The bacteria were inoculated in MRS broth (10 ml), then incubated at 37°C for 48 h. The viability of L. acidophilus cells was evaluated by total plate counting and MRS agar and plate count agar used for L. acidophilus counting28.
Sample preparation
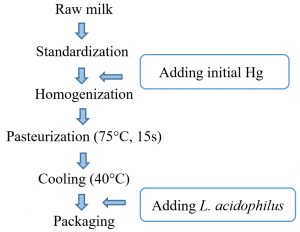
The milk samples were designed according to the following schematic diagram (Fig. 1). Then the analysis was carried out through storage period.
Hg Analysis
The inductively coupled plasma mass spectrometer (ICP- MS) 4500a (England) was applied in this study, with a cross flow rate nebulizer and a Peltier-cooled quartz spray chamber. It was tuned up by an aqueous multi-element before each experiment. At first all the prepared samples were under digestion by using the microwave with segmented rotor MPR-600 (using pressure up to 35 bar; at 260°C) 29.
Plackett-Burman Design and selection of variables
The bacterial concentration, inoculation temperature, contact time, Mercury concentration and shaking rate are the effective independent variables on Mercury bioremoval by L. acidophilus as mentioned in previous studies6, 30. The variables were selected in this project by the help of literature reviews and pre-experience study (Table 1). Table 1 shows PBD for evaluation of 5 process variables in two levels.
Table (1):
Plackett-Burman for evaluation of impact of the variable on Mercury biosorption by L. acidophilus.
Run |
Bacterial concentration (CFU) |
Inoculation Temperature (°C) |
Contact time (day) |
Mercury concentration (μg/L) |
Shaking rate (rpm) |
|---|---|---|---|---|---|
1 2 3 4 5 6 7 8 |
1×1011 1×1012 1×1011 1×1012 1×1011 1×1012 1×1011 1×1012 |
40 4 4 40 4 4 40 40 |
4 1 4 1 1 4 1 4 |
100 100 100 100 40 40 40 40 |
0 50 50 0 0 0 50 50 |
The levels of Mercury concentration were selected by the aim of this project to study the potentiality of L. acidophilus to remove the low levels of Mercury (μg/L) in milk. Up to now there is no published information on this issue. For designed experiment of biosorption, the bacterial concentration (1 ×1011 and 1×1012 CFU) was added to sterile milk of 37°C in 250 mL Erlenmeyer flasks with the rest time of 20 minutes then Mercury (40 and 80 μg/L) added to the flasks. After that the flasks were put at on the shaker. At the contact time (1st or 4th day), bacteria cells were centrifuged at 8000 × g for 20 min. Then the supernatant was analyzed for residual Mercury concentration by ICP- MS. All these experiments were carried out in triplicates. The ability of L. acidophilus to absorb Mercury was estimated by the following equation31:
% Removal= 100 × [(C0 – C1) / C0]
Where C0 is the initial and C1 is residual Mercury concentration.
The data were analyzed using the Minitab (version 14) statistical software. According to the variance analysis, the 3 main variables were: Mercury concentration, L. acidophilus concentration and the contact time.
Response surface methodology (RSM)
RSM is the usage of statistical and mathematical techniques together for analyzing the independent variables on the responses. RSM is a helpful application in optimizing and improving the precess design. This method is more practical by applying interactive computer programs between the variables based on experimenter’s prior knowledge. After all it represents the parameters effects on the process32.
The Plackett-Burman results showed that 3 variables; L. acidophilus concentration, Mercury concentration and contact time, having significant effect on mercury bioremoval. RSM was designed for a completed determination of optimum variable levels for Mercury bioremoval and also elimination the tests number. In our project, CCD was used to find the optimal bioremoval conditions with the experimental factors levels displayed in the Table 2.
Table (2):
Main variables and levels for Mercury biosorption by L. acidophilus by central composite design.
| Range and level | |||||
|---|---|---|---|---|---|
| Independent process variable | − α (−1.68) | -1 | 0 | +1 | + α (+1.68) |
| L. acidophilus concentration (CFU) | 1× 1010 | 10×1011 | 1×1012 | 10×1013 | 10× 1014 |
| Initial Hg concentration) μg/L) | 40 | 50 | 70 | 90 | 100 |
| Contact time (day) | 0 | 1 | 2 | 3 | 4 |
The other factors were kept constant as the following: the inoculation temperature at 25°C and the shaking rate at 50 rpm. For data analysis the Design- Expert 7.1.5 (Stat-Ease Inc., USA) software was used. The bioremoval runs were performed by the 5 levels of each variable (Table 2) and then the tests obtained by CCD.
Evaluation of the capacity of binding metal
The maximum capacity of binding Mercury would be predicted with the different isotherm models like Langmuir and Freundlich. The absorption models used for explaining absorption system of the bacterial cells in biosorption of metals at the contact time33,34.
Statistical Analysis
The results of statistical analysis were done by MINITAB statistical software (version 14) and the response surface plots were prepared. The statistical data were provided by analysis of variance. All data are represented as the mean value ± standard deviation (M ± SD) of independent experiments in mentioned days. P-values below 0.05 were statistically significant.
RSM for optimization of Mercury bioremoval
The analysis of variance showed the effect of the variables designed by Plackett-Burman Design. Using RSM after analysis of variance showed that the Mercury bioremoval level is the result of the 3 variables shown in Table 3. The P-values <0.05 showed that the model terms are significant. In this study Mercury concentration, contact time and biomass dosage are significant model terms.
Table (3):
Analysis of variance of parameters studied for Mercury biosorption by L. acidophilus.
Source |
Sum of Squares |
df |
Mean Square |
F-value |
p-value |
|---|---|---|---|---|---|
Shaking rate |
0.211 |
1 |
0.211 |
2.88 |
0.4125 |
Mercury concentration |
9.87 |
1 |
9.87 |
94.02 |
0.0542 |
L. acidophilus concentration |
9.38 |
1 |
9.38 |
78.35 |
0.0712 |
Contact time |
9.80 |
1 |
9.80 |
82.99 |
0.0688 |
Inoculation temperature |
0.382 |
1 |
0.382 |
3.42 |
0.3202 |
Study the influencing factors on the effect of L. acidophilus on Mercury bioremoval
The factors influencing on biosorption of mercury by L. acidophilus in milk were analyzed and described below:
Effect of L. acidophilus concentration and contact time on removal efficiency
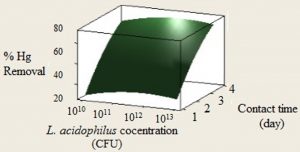
In this study, the experiments were done for evaluating the ability of L. acidophilus concentration in the range of 1010 to 1013 CFU on Mercury biosorption efficiency during the contact times of 1 to 4 days. Fig. 1. shows the data collected from Mercury biosorption by L. acidophilus at different contact times. As it shows the maximum binding rate of Hg occurred in the 4th day. The removal efficiency of this heavy metal first enhanced with rising the bacterial concentration and contact time and reached to the maximum level and the further increase of bacterial concentration, caused a light decrease in removal level.
In general, heavy metals biosorption is a complicated mechanism. There are 3 theories in metal binding; the ion exchange with cell walls’ teichoic acid and peptidoglycan, the precipitation and the ligands formation20.
Lactic acid bacteria are gram positive and their cell walls contain a thick layer of teichoic acid, peptidoglycan and exopolysaccharides. The surface functional groups; carboxyl, hydroxyl, phosphate make the negative charges in L. acidophilus. So, the bacteria would be able to absorb the cationic ions of heavy metals20,35.
The light decrease trend in removal process could be explain as a result of the bacteria partial aggregation at higher concentrations that causes the decrease in free sites in surface protein and exopolysaccharides and finally decreased biosorption35,36. In this study, the highest Mercury removal efficiency was 72% in biomass of 1×1012 CFU in the 3rd day. Similar studies reported the same results for increasing biosorption during exposure time by Halttunen35 for Lactic acid bacteria, Rayes37 for L. rhamnosus and L. fermentum. It has been reported that metal binding is a process carried out on the bacteria cell surface efficiently with no energy consumption35.
Fig. 2. Contour plot showing interactive effect of L. acidophilus concentration dosage and contact time on the Mercury removal
As shown in Fig. 2. Mercury bioremoval enhanced by increasing the contact time from 1st to 4th day in addition to rising the bacteria concentration. The optimum level of L. acidophilus was 1×1012 CFU. It is sensible that by increasing the contact time, more Mercury ions would be connected to bacteria surface receptors and the bioremoval process would be more efficient by time.
Effect of L. acidophilus concentration and Mercury concentration on removal efficiency
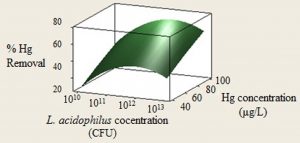
The effect of L. acidophilus concentration and initial Mercury concentration on the biosorption was investigated in the range of 1010 to 1013 CFU and 40 – 100 μg/L, respectively (Fig. 3.). The results revealed that by increasing the Mercury concentration, the adsorption increased. As shown in Fig. 3. Increasing L. acidophilus concentration up to 1×1012 CFU, make the removal efficiency enhancing. The maximum Mercury removal efficiency (72%) was observed at the initial Mercury concentration of 80 μg/L and the biomass concentration of 1×1012 CFU. L. acidophilus shows a high affiliation for biosorption of heavy metals35,38. Heavy metals binding is a surface process as the presence of anionic functional groups and it also depends on the capacity of the bacteria strains and the metal electronegativity20. It is reported that metal absorption in bacteria cells could be explained by the interactions between the heavy metals and the negative charge of bacteria surface. Gram positive bacteria like L. acidophilus have some polymers like lipoteichoic acid in their cell wall that can be responsible for such interactions20,39.
Fig. 3. Contour plot showing interactive effect of L. acidophilus concentration and Hg concentration on the Mercury removal
It is observed that by rising the metal concentration, the absorption to the bacteria receptors would also increase, which results in the higher bioremoval level38,40.
According to our findings Mercury biosorption efficiency increased by increasing the Mercury concentration in the range of 40 to 100 μg/L. The important factors as shown in Fig. 3, are Mercury concentration and the L. acidophilus concentration for Mercury bioremoval and their optimum levels are 80 μg/L and 1×1012 CFU for the maximum level (72%) of the biosorption. The same results were reported by Dobrowolski40, Allam41, Akhmetsadykova42 and Halttunen35 as the absortion would improve by increasing the bacterial concentration. Also by increasing the metal concentration, the biosorption would enhance as mentioned in some studies by Massoud18 Halttunen35, Shameer36, Kinosita38.
Isotherm model studies
The capacity of L. acidophilus concentration (1012 CFU/mL) for Mercury bioremoval was determined at different mercury initial concentrations (20, 40, 60, 80 and 100 μg/L). The biosorption isotherms are detemined by using the isotherm models such as Langmuir and Freundlich. The regression coefficient (R2) show the best isotherm describing the Mercury biosorption by L. acidophilus. All the experiments were performed in three replications.
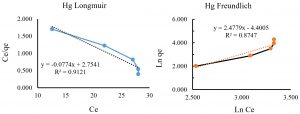
The Langmuir model is the most common model used in scientific studies. The Langmuir equation is correct for monolayer absorption using the following equation33:
Ce / Qe = 1/ (K * Qmax) + Ce / Qmax
Where Qe (µg/L) is the amount of Hg in absorbing equilibrium, Ce (µg/L) is the equilibrium concentration of Hg in milk, Qmax (µg/L) is the maximum Hg absorption in high Ce level; and KL (L/µg) is the Langmuir constant. The Ce/Qe versus Ce indicate a straight line of slope 1/Qmax and also intercept of 1/KLQmax.
The Freundlich equation is as the following equation34:
Ln Qe = Ln Kf + 1/n Ln Ce
Where Kf and n is the Freundlich constants. The parameters KF and n is defined from the linear plot of ln Qe versus ln Ce. Freundlich equation varies with the materials heterogeneity.
The Langmuir and Freundlich models’ parameters are given in Table 4.
Table (4):
Langmuir and Freundlich isotherm parameters of Hg in various initial concentrations.
| Hg initial (µg/L) |
Langmuir model | Freundlich model | |||
|---|---|---|---|---|---|
| Ce | Qe | Ce/Qe | Ln Qe | Ln Ce | |
| 20 | 10.5 | 9.4 | 1.128 | 2.241 | 2.361 |
| 40 | 18 | 22 | 0.818 | 3.091 | 2.890 |
| 60 | 22 | 38 | 0.563 | 3.648 | 3.073 |
| 80 | 24 | 56 | 0.429 | 4.025 | 3.178 |
| 100 | 25 | 75 | 0.333 | 4.317 | 3.219 |
As shown in Fig. 4. A and B, the biosorption enhanced by increasing the initial Mercury concentration, because more metal concentration supplied more possible ions of Mercury ions to bind with absorbents’ functional groups6,43. By comparing the both R2 values in Langmuir and Freundlich models, it was inferred that Langmuir isotherm model showed better fit than Freundlich model, which also confirm that Freundlich equation is correct for monolayer absorption on surface binding. The higher correlation coefficient in Langmuir model indicates the Mercury absorptions obey Langmuir isotherm model.
The mercury presence in water and food is a public health problem. The European Rapid Alert System for Food and Feed (2018)44 have reported that heavy metals are the contaminants that attracts the high notifications in water and food and Lead, Cadmium and Mercury are the ones that make the most problems for people. Among all the food and drinks, milk is the most sensible one that should be safe enough to be consumed.
In this project, RSM was used to evaluate the optimal condition for Mercury bioremoval by of L. acidophilus. Our findings showed the highest level of Mercury bioremoval of 72% in the concentration of 1×1012 CFU, the Mercury concentration of 80 μg/L and in the 4th day. The biosorption increased by increasing the metal and bacteria concentration as well as the contact time. This study represented the ability of L. acidophilus for Mercury removal in very low concentration levels (µg/L) from milk. Also, these findings open the doors of investigating the capacity of Mercury binding by LABs in milk. Further studies are suggested for other LAB strains in milk and foodstuffs to reduce the toxic effects of the heavy metals.
ACKNOWLEDGMENTS
We would like to thank the National Nutrition and Food Technology Research Institute (NNFTRI) of Iran for supporting this project. The authors would like to thank Dr. A. Zoghi and for her kind assistance.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors have made substantial contribution to the work and approved it for publication.
FUNDING
This work was supported by Shahid Beheshti University of Medical Sciences (Grant number 22408).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
Not applicable.
- Arif TJ, Mudsser A, Kehkashan S, et al. Heavy Metals and Human Health, Mechanistic Insight into Toxicity and Counter Defense System of Antioxidants. Int J Mol Sci. 2015;16(12):592-630.
Crossref - Jaiswal G, Singh R, Porwal S. Bioremediation of Mercury through Encapsulation of the Clone Carrying meroperon. J Pure Appl Microbiol. 2019;13(1):553-560.
Crossref - Wang Y, Dang F, Zhong H. Effects of sulfate and selenite on mercury methylation in a mercury-contaminated rice paddy soil under anoxic conditions. Environ Sci Pollut Res. 2016;23:4602-4608.
Crossref - McConnell JR, Edwards R. Coal burning leaves toxic heavy metal legacy in the Arctic. Proc Natl Acad Sci. 2008; 105(34):12140-12144.
Crossref - Lucey JA. Raw Milk Consumption, Risks and Benefits. Nutr Today. 2015;50(4):189-193.
Crossref - Jadan-Piedra C, Alcantara C, Monedero V, Zuniga M, Velez D, Devesa V. Effect of lactic acid bacteria on mercury toxicokinetics. Food Chem Toxicol. 2009;128:147-153.
Crossref - Bjorklunda G, Dadarb M, Mutterc J, Aaseth J. The toxicology of mercury, Current research and emerging trends. J Environ Res. 2017;159: 545-554.
Crossref - Codex stan 193. Codex General Standard for Contaminants and Toxins in Food and Feed. 2009.
- Wang MQ, Wang ZT, Bao DY, Ran Chin L. Food contamination monitoring and analysis in 2000 in China. J Food Hyg. 2002;20:185-200.
- Najarnezhad V, Akbarabadi M. Heavy metals in raw cow and ewe milk from north-east Iran. Food Add Contamin. 2013;3:158-162.
Crossref - Naushad M, Vasudevan S, Sharma G, Kumar A, ALOthman ZA. Adsorption kinetics, isotherms, and thermodynamic studies for Hg2+ adsorption from aqueous medium using alizarin red-S-loaded amberlite IRA-400 resin. Desalin Water Treat. 2016;57(39): 18551-18559.
Crossref - Naushad M., ALOthman ZA, Awual MR. Adsorption kinetics, isotherms, and thermodynamic studies for the adsorption of Pb2+ and Hg2+ metal ions from aqueous medium using Ti(IV) iodovanadate cation exchanger. Ionics, 2015;21(8):2237-2245.
Crossref - Awual R, Hasan M, Eldesokyc G, Khaleque A, Rahman MM, Naushad M. Facile mercury detection and removal from aqueous media involving ligand impregnated conjugate nanomaterials. Chem Engine J. 2016;290:243-251.
Crossref - Massoud R, Hadiani MR, Khosravi Darani K. Bioremediation of heavy metals in food industry Application of Saccharomyces cerevisiae. Electron J Biotechnol. 2019a;37:56-6.
Crossref - Chen W, Narbad A. Lactic Acid Bacteria in Foodborne Hazards Reduction The use of lactic acid bacteria to reduce mercury bioaccessibility. Springer Nature, Singapore. 2018.
Crossref - Eman Abdullah MA, Mohsen AS, Tahany MA, Abdel-Rahman AM. Bioremediation of Waste Water from Cadmium Pollution using Silicon Dioxide Nanoparticles and Fungal Biomasses. J Pure Appl Microbiol. 2019;13(3):1561-1570.
Crossref - Hadiani MR, Khosravi-Darani K, Rahimifard N, Younesi H. Assessment of Mercury biosorption by Saccharomyces cerevisiae, Response surface methodology for optimization of low Hg (II) concentrations. J Environ Chem Engin. 2018;6(4):4980-4987.
Crossref - Massoud R, Khosravi-Darani K, Sharifan A, Asadi GH. Lead bioremoval from milk by Saccharomyces cerevisiae. Biocatal Agric Biotechnol. 2019b;22:11-20.
Crossref - Massoud R, Khosravi-Darani K, Sharifan A, Asadi, GH. Cadmium Bioremoval by Saccharomyces cerevisiae In Milk. J Med Microbiol Infect Dis. 2020;12:22-30.
Crossref - Zoghi A, Khosravi-Darani K, Sohrabvandi S. Surface binding of toxins and heavy metals by probiotics. Mini Rev Med Chem. 2014;14:84-98.
Crossref - Mrvcic J, Stanzer D, Solic E, Stehlik-Tomas V. Interaction of lactic acid bacteria with metal ions: opportunities for improving food safety and quality. World J Microbiol Biotechnol. 2012;28(9):2771-2782.
Crossref - Tan H, Wang C, Zeng G, Luo Y, Li H, Xu H. Bioreduction and biosorption of Cr(VI) by a novel Bacillus sp. CRB-B1 strain. J Hazard Mat. 2020;386:12-18.
Crossref - Shenghui X, Ruixiang X, Zhongren N, Peng C. Bioadsorption of arsenic from aqueous solution by the extremophilic bacterium Acidithiobacillus ferrooxidans DLC-5. Biocat Biotrans. 2018;37(1):35-43.
Crossref - Majlesi M, Shekarforoush SS, Ghaisari HR, Nazifi S, Sajedianfard J. Effect of Bacillus coagulans and Lactobacillus plantarum as probiotic on decreased absorption of cadmium in rat. J Food Hygin. 2017;6(22):25-33.
- Hirak R, Dash S, Das S. Corrigendum to “Bioremediation of inorganic mercury through volatilization and biosorption by transgenic Bacillus cereus BW-03. Int Biodet Biodeg. 2015;103:179-185.
Crossref - Beladi M, Akhavan Sepahi A, Mehrabian S, Esmaeili A, Sharifnia F. Antibacterial Activities of Selenium and Selenium Nano-particles from Lactobacillus acidophilus on Nosocomial Strains Resistant to Antibiotics. J Pure Appl Microbiol. 2015;9(4):2843-2851.
- Singh AL, Sarma PN. Removal of arsenic from water using Lactobacillus acidophilus. Biorem J. 2010;14(2):92-97.
Crossref - Vinderola CG, Reinheimer JA. Enumeration of Lactobacillus casei in the presence of Lactobacillus acidophilus Bifidobacteria and lactic starter bacteria in fermented dairy products. Int Dairy J. 2000;10(4):271-275.
Crossref - Khan N, Jeong S, Hwang M, et al. Analysis of minor and trace elements in milk and yogurts by inductively coupled plasma-mass sperometry (ICP-MS). Food Chem. 2014;147:220-224.
Crossref - Pakdel M, Soleimanian-Zad S, Akbari-Alavijeh S. Screening of Lactic acid bacteria to detect potent biosorbents of lead and cadmium. Food Control. 2019;100:144-150.
Crossref - Goksungur Y, Uren S, Guvenc U. Biosorption of cadmium and lead ions by ethanol treated waste baker’s yeast biomass. Biores Technol. 2015;96:103-109.
- Bas D, Boyac IH. Modeling and optimization usability of response surface methodology. J Food Eng. 2007;78(3):836-845.
Crossref - Langmuir I. The adsorption of gases on plane surfaces of glass mica and platinum. J Am Chem Soc. 1918;40(9):1361-1403.
Crossref - Freundlich HM. The adsorption in solutions. Chem. 1906;57:385-470.
- Halttunen T, Salminen S, Jussi M, Raija T, Kalle L. Reversible surface binding of cadmium and lead by lactic acid and bifidobacteria. Int J Food Microbiol. 2008;125(2):170-175.
Crossref - Shameer S. Biosorption of lead copper and cadmium using the extracellular polysaccharides (EPS) of Bacillus sp. from solar salterns. 3 Biotech. 2016;6(2):194 – 200.
Crossref - Rayes AAH. Field studies on the removal of lead cadmium and copper by the use of probiotic lactic acid bacteria from the water for culturing marine tilapia T. spilurus. New York Sci J. 2012;5(11):120-125.
- Kinoshita H, Yui S, Fumika O, et al. Biosorption of heavy metals by lactic acid bacteria and identification of mercury binding protein. Res in Microbiol. 2013;164(7):701-709.
Crossref - Alcantara C, Jadan-Piedra C, Velez D, Devesa V, Zuniga M, Monedero V. Characterization of the binding capacity of mercurial species in Lactobacillus strains. J Sci Food Agricul. 2017;97(15):5107-5113.
Crossref - Dobrowolski R, Szczes A, Czemierska M, Jarosz-Wikolazka A. Studies of cadmium (II) lead (II) nickel(II) cobalt(II) and chromium(VI) sorption on extracellular polymeric substances produced by Rhodococcus opacus and Rhodococcus rhodochrous. Biores Technol. 2017;225:113-120.
Crossref - Allam NG, Ali EM, Samya S, Abd-Elrahman E. The role of probiotic bacteria in removal of heavy metals. Egyp. J Environ Res. 2015;3:1-11.
- Akhmetsadykova S, Konuspayeva G, Loiseau G, et al. Protection against lead contamination by strains of lactic acid bacteria from fermented camel milk. J. Food Agricul. 2013;25(4):274-282.
- Mironyuk I, Tatarchuk T, Naushad M, Vasylyeva H, Mykytyn I. Highly efficient adsorption of strontium ions by carbonated mesoporous TiO2. J Molecul Liquids. 2019;285:742-753.
Crossref - European Rapid Alert System for Food and Feed. European Commission annual report. 2018.
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.