Heavy metal contamination from anthropogenic activities has an adverse effect on the environment due to its cytotoxicity, carcinogenicity and mutagenicity. Environment harbours microorganisms, some of which have been found to modify physico-chemical conditions of their surrounding environment through certain processes such as detoxification, metal homeostasis, precipitation, redox transformations etc. Investigations in the past have shown that short term contact with metals of certain bacteria causes the selection of resistant bacteria within weeks, while a prolonged exposure showed selected strains able to thrive better. Hence biotic methods could assist removal of heavy metals based on biosorption or bioaccumulation by microorganisms, which are cost-effective and environmental friendly in the long run. Microbial remediation is influenced by biotic and environmental factors as also the contamination site characteristics. The aim of this paper is to highlight and review some of the mechanisms of microbial remediation through techniques such as biostimulation, bioaugmentation etc.
Heavy Metals, Biosorbents, Microbial Remediation
Microorganisms are widespread in all the realms of the biosphere growing in an extensive range of environmental conditions. The nutritional versatility of different microbes is being explored in the area of biodegradation of pollutants. Although industrialization and technological advancement in today’s times is inevitable, multiple ecological and human catastrophes have continuously occurred over the past few decades that have resulted in the industries playing a huge anthropogenic factor for various reported environmental pollution episodes. Industrial discharges specifically impact the lotic water quality with the presence of toxic and hazardous persistent chemicals like lead, cadmium and mercury and toxic organic chemicals such as pesticides, PCBs, dioxins, polyaromatic hydrocarbons (PAHs), etc. which have a deleterious effect on human health.1
The persistence of heavy metals in the environment and their toxic nature cause bioaccumulation thereby causing deleterious effects on organisms with an exposure to metals with high concentrations when enzymes, cellular proteins and nucleic acids react with them to form complexes.2
Toxic heavy metals have a tendency to bioconcentrate, bioaccumulate and thereby biomagnify thus causing ecological risks. Trophic transfers of four common toxic metals namely Cd, Cr, Cu and Hg investigated in the food webs in the Bohai Sea of North China exhibited Hg and Cr to be biomagnified between the trophic levels while Cu underwent biodilution and Cd exhibited no tendency to biomagnify though its presence in one species was shown to exceed the restrictive criteria which if consumed by another organism could pose sufficient ecological risks.3
Fish being a primary source of nourishment in many parts of the world, any bioaccumulation of toxic heavy metals in fish is of concern.4 High levels of heavy metals in fish may lead to toxicity in humans.5 Consumption of any fish growing in industrial effluent contaminated water bodies poses high human health risks.6 Likewise, heavy metal-contaminated crops if consumed as food could also lead to significant risks to human health.7 Agricultural fields if irrigated with sewage wastewater may cause heavy metals to accumulate in soils and eventually they get transferred to food crops such as cereals and vegetables as well as to milk.8 Heavy metals tend to move from the abiotic to the biotic environment, accumulating in organisms at different trophic levels of the food chain or food web. Trophic transfer, bioaccumulation and biomagnifications of hazardous heavy metals in the nutrition network have important implications on wildlife and human health.9
Some heavy metals like Cr, Cu, As, Cd, Hg, Pb, Ni, Se, Zn, Ag etc. have also been reported to be toxic at low concentrations due to their cytotoxicity, carcinogenicity and mutagenicity.10 Studies conducted have also demonstrated that oxidative stress and reactive oxygen species (ROS) production are influenced by the toxicity and carcinogenicity of some heavy metals such as Cd, Cr, As, Pb and Hg, those that affect public health because of their toxic nature even in minor quantities.11
Interactions of Microorganisms with Heavy Metals
Functional groups in the biomolecules of the cell are obstructed when excessive concentrations of metals may displace the enzyme metal ions and participate in cell reactions by competing with structurally related non-metals.12 Metals in combination may have more negative impacts on the diversity of the microbial biomass in comparison to the high concentrations of single metals,13 even affecting the physiological functions in organisms when these form complex toxic compounds.14
Studies on certain soil bacteria have demonstrated that selection and growth of resistant bacteria can occur within weeks of a short term exposure of bacterial strains to heavy metals but only specific strains of resistant bacteria grow when exposed to metals for a longer duration.15 Some general effects of toxicity of heavy metals on microorganisms are listed in Table 1.
Table (1):
Toxicity of heavy metals to microorganisms.
Heavy Metals |
Sources |
Effects on microbes |
Citation |
|---|---|---|---|
As |
Contaminated soils, sediments, groundwater |
Enzyme deactivation |
[16, 17] |
Cd |
Sedimentary rocks, Anthropogenic inputs |
Protein denaturation, nucleic acid damage, affects cell division and transcription |
[16,18] |
Cr |
Anthropogenic sources |
Growth suppression, inhibition of oxygen absorption |
[16,19] |
Cu |
Rocks and soil |
Cellular function disruption, inhibition of enzyme activities |
[16, 20] |
Se |
Minerals |
Hinders growth rate |
[16, 21] |
Pb |
Anthropogenic sources |
Protein/nucleic acid destruction, inhibition of enzyme action and transcription |
[16, 20] |
Hg |
Natural sources and used in industrial applications |
Protein denaturation, inhibition of enzyme activity, disrupts plasma membrane |
[16, 20] |
Ni |
Sulphide ores and anthropogenic sources |
Disrupts cell membrane, hinders enzyme functions and cause oxidative stress |
[16, 20, 22] |
Ag |
Ores |
Cell lysis, inhibition of cell growth |
[16, 23] |
Zn |
Soil and sediments |
Death and decline in biomass, inhibition of growth |
[16, 22] |
Cr and Cd have the ability to weaken the bioremediation capacity and denaturing microbial flora.16 Mutagenesis is found to be caused within the cell due to transcription and replication being affected when Cr (III) reacts with carboxyl and thiol groups thus changing the enzymatic structure and the cationic Cr (III) complexes formed within the cell interact with phosphate groups of DNA which are anionic.19 Superoxide radicals that tend to damage the DNA structure are stabilized when in contact with Al.23 Enzyme configurations could be altered through the allosteric effects by heavy metals that go into competitive or noncompetitive interactions with substrates thus affecting the vital enzymatic functions.24 Ligand interactions and metal displacements from native binding sites in the presence of Cd and Pb cause damage to the DNA structure and cell membranes of the microorganism.25 Microbial growth, metabolism and morphology are affected by a change in the nucleic acid structure with functional disturbances being caused thus affecting the physiology and disrupting the cell membranes and thereby inhibiting enzyme activity.20
Microbial Remediation of Heavy Metals
Interactions of Bacillus subtilis with multiple metals like Cu, Fe, Mg, Au and Pb were studied in the early 1980s,26-28 wherein it was found that differences between the net anionic charge of the bacteria and the net positive charge of metals were what led to an interaction of the bacterial cell to the metals. The metals with the opposite charges were able to bind to the cell surface of the microbes on the nucleation sites which eventually caused precipitation of the metals on the cell wall.29 Studies by the U.S. Geological Survey (USGS) revealed that interactions of certain species of bacteria and fungi with both metals and other toxic compounds led to the concept of bioremediation being explored further.30, 31
USGS in 1992 explored the possibility of activating bacterial species in the soil by the addition of certain nutrients to the contaminated soils in Hanahan, SC.30-32 Observations revealed that removal of about 75% of the toxic chemicals in the soil was effected within a span of a year. The utilization of natural microorganisms found in soil, water and sludge to eradicate pollutants thus spearheaded the study and further applications of bioremediation in the environment.30 Even the genetically engineered microorganisms (GEM) were found to be capable of degrading the environmental toxins and bound metals as was found by the study conducted in the University of Tennessee and Oak Ridge National Laboratory wherein the genetically engineered microorganism Pseudomonas fluorescens (KH44) designed in the laboratory was found to degrade toxic polycyclic aromatic hydrocarbons.30, 33
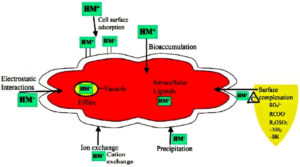
Microbial cells through interactions can adsorb or absorb the heavy metals onto the binding sites on the cellular surface21 depending on the kinetic equilibrium and metal composition at the surface of the cell. Biosorption is thus affected through electrostatic interactions, redox processes, ion exchange, precipitation and surface complexation.34 Cellular fragments, dead or live biomass can carry out biosorption through passive uptake via surface complexation (Figure 1) on to the cell wall and other outer layers. This process is not influenced by cell metabolic cycle.36
During an active uptake, the heavy metal ions pass across the plasma membrane into the cytoplasm (method of bioaccumulation) during which the cell is influenced by physical, chemical, and biological processes including intracellular and extracellular processes.36, 37 An organism capable of bioaccumulation must be able to transform the metal, changing it from a toxic to a harmless form even when tolerating the high concentrations of one metal or a combination of metals. The surface structures of bacteria play a vital role in how the bacteria interacts with the surrounding environment. While the cell walls of the gram-negative bacteria contain lipopolysaccharide, phospholipids and peptidoglycan layers, those of the gram-positive bacteria have minor amounts of teichoic acid usually present along with the peptidoglycan in several layers, the latter forming as much as 90 % of the cell wall.38, 39
Bioremediation in Industrial Waste Treatment
Through bioremediation, industrial waste and wastewater treatment have been successfully adopted for multiple years. Waste treatment plants are known to employ specialized microbial populations in the treatment of industrial effluents. Microorganisms have been found to enzymatically metabolise many xenobiotics thereby degrading environmental contaminants.40
A review done on the feasibility of microplastics removal through water treatment from water sources reveals that sludge and membranes carry more microplastics from water treatment. Primary treatment in a wastewater treatment plant was reported to remove 16.5 to 98.4% microplastics while secondary treatment showed a removal efficiency ranging from 78.1 to 100% with the tertiary treatment showing a removal of 87.3% to above 99.9% microplastics. It was however reported that a complete elimination of microplastics from the final treatment plant wastes was not feasible since the wastes removed from the processes get disposed back to the environment in the form of sludge and disposed membranes.41
Marine enzymes are successfully used as an alternative in terms of some kinds of industrial waste treatment. Industrial pollutants as those from metal smelting, petrochemical waste, mining wastes, paper and pulp industry wastes, chemical weapon producing industry wastes, wastes from dye industry, anthropogenic activity and agriculture tend to pollute the marine environment.42 Isolates of bacteria and fungi are found to remediate the dye effluents.43 Partially degraded lignin and chlorinated phenolic compounds are sources of pollutants released into the environment from paper and pulp industries. These wastes are bioremediated with fungal species that produce extracellular enzymes (oxidoreductase), viz., manganese peroxidases, lignin peroxidases, and laccase from fungal mycelium, being a quicker bioremediation process than bacteria.44 Generally, enzymes are more active than microorganisms to utilize the substrate and its transfer from complex to simple form or smaller substances.45 Most of the enzymes are ecologically beneficial and risk-free for the environment, being able to play a vital role in the bioremediation of adverse compounds in the environment.
Mechanism of Detoxification of Heavy Metal by Microbes
Degradation, removal, alteration, immobilization or detoxification of chemical and physical wastes through the action of microbial flora in the environment is the process of bioremediation, a technology that could also exploit naturally occurring mitigation measures where no human intervention is required other than monitoring as is the case of natural attenuation which relies only on the natural conditions.46
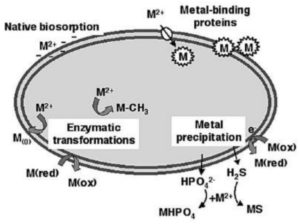
Degradation of a pollutant is encouraged when microorganisms play the role of catalysts by facilitating the progress of biochemical reactions that decompose the desired pollutant chemical through various enzymatic pathways.46 The physical and chemical conditions of the immediate environment of the microorganisms are modified through processes like detoxification and redox transformations.47, 48 The processes of microbial interactions with metals and their locations in the bacterial cell are shown in Figure 2.
Figure 2. Bacterial interaction with heavy metal ions (M2+) in the environment, with reference to the cellular compartment where bacteria response happens. [49]
In bioaccumulation, the uptake mechanisms of metal into the bacterial cell have been studied to be through processes as passive diffusion, facilitated diffusion and active transport.50 Heavy metal polluted environments, specifically soil and water have been bioremediated through the use of Cupriavidus metallidurans CH34 which has the capacity to bioconcentrate Se and Au and to volatilize Hg through multiple reactions.39, 51, 52 Pseudomonas stutzeri, a microorganism isolated from a foundry soil demonstrated good resistance to the toxicity of Cr to a concentration of up to 1mM. It was also found to reduce Cr (VI) up to 100 µM anaerobically.53
Processes such as ion exchange, adsorption, and covalent bonding that occur due to the chemiosmotic gradient across the cell without the use of ATP are found to be the crux of biosorption.54, 55 Biosorption mechanisms are formulated on two factors; dependence on cell metabolism and area inside the cell from where the metal is eliminated.56
Biostimulation is when the activities of indigenous microorganisms are stimulated through the use of specific nutrients into the soil or water so as to encourage the removal of the metal from the site.46 Application of plant nutrients, growth supplements and essential trace elements as also changes made to the conditions like pH, oxygen and temperature will help accelerate the rate of metabolic reactions and the metabolic pathways,57, 58 and thus stimulate the naturally existing bacterial and fungal communities. The population of microorganisms in contaminated soil also needs to be in large numbers so as to bring about effective bioremediation. Small concentrations of pollutant can stimulate the microorganisms by activating the operons for the bioremediation enzymes. The use of nutrients like N, P and C will encourage the microorganisms to create the basic environment needed for pollutant degradation.46, 59
Large array of physical, chemical and biological processes that contribute to the eradication of the pollutants from surroundings through processes such as biodegradation (aerobic and anaerobic), uptake of chemicals by biota, physical phenomena such as diffusion, dispersion, dilution, sorption/desorption, volatilization and reactions such as ion exchange and complexation is the basis of bioattenuation,60 which ultimately help reduce the toxicity, mass or volume and concentration of contaminants in the environment.
Bioaugmentation is a method of biodegradation wherein the biodegradable microorganisms, be it engineered, exotic or natural, are introduced into the environment to aid in natural microorganism population growth to increase the capacity of biodegradation of the native microbial populations thus effecting enhanced degradation through preferential feeding of the metal contaminants on site. Microorganisms collected from the contaminated site can be cultured or genetically modified before being introduced back to the site.46 Through this process, even complex pollutants can be eliminated quickly.60
Studies conducted using crude oil as a growth substrate showed that microbial consortia from oil- contaminated sites efficiently degraded complex hydrocarbons.61 The degradative efficiency is more enhanced in the genetically modified microorganisms due to diverse metabolic profiles that can aid it in biodegradation.62, 63 Research has shown that effective degradation of a wider range of chemical and physical environmental pollutants has been possible using genetically engineered microorganisms in bioremediating soil, groundwater and activated sludge.60, 64
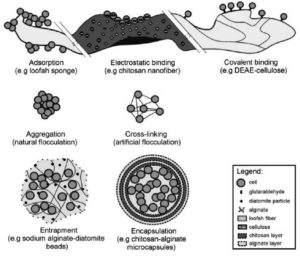
Mobility of microbial cells or enzymes can be arrested by adopting techniques by still preserving its viability and catalytic functions through the process of immobilization.65-70 This brings about numerous benefits like greater degradative efficiency of pollutants, less probability of alterations in the gene, resistance of biocatalysts to changes in the immediate environment and tolerance of the biocatalysts to the excessive pollutant concentrations.65, 69-72
Some techniques of immobilization65, 70,72 identified have been adsorption, electrostatic or covalent binding on the surface, flocculation, encapsulation and entrapment (Figure 3). Immobilization of the cells and enzymes of microorganisms through the technique of adsorption is achieved through physical interaction with the external surface of water-insoluble carrier molecules employed in bioremediation forming weak bonds.70 In electrostatic binding, the water insoluble carriers’ surfaces are washed with a buffer solution. This makes the surface hydrophilic that promotes the attraction of negatively charged cells or enzymes.70,73,74 With the technique of entrapment in bioremediation, microorganisms are entrapped in a porous matrix thus restricting their mobility within the carrier. The advantage with this is that there is limited exchange of nutrients and metabolites and thus no leaks from the system to the environment. The entrapped microbes are physiologically varied with the cells closer to the surface showing more metabolic activity as opposed to the starved cells seen in the inside of the heterogenous carrier.65,69,70,72,75 Entrapment is a quick, nontoxic, economical method. The entrapped microorganisms are guarded against any change in the environmental factors.69, 70 The use of a semi-permeable membrane to set apart the immobilized particles from the external environment is the process termed as encapsulation.70 Though the microorganisms are significantly protected against the unfavourable conditions of the external environment, their use in ex- situ bioremediation is limited due to the probable damage that could be caused to the growing cells.65, 70, 72, 76
With the introduction of selected reagents into metal-polluted water, the redox reactions involved modifies the chemistry of the water on-site thus enhancing the decomposition and extraction of various contaminants.56, 77 Studies have shown that heavy metals with high toxicity like As, Cr, Hg and Se in soils or sediments have also been transformed into innocuous forms with less toxicity through the oxidation-reduction reactions happening in situ.78-80 Oxidation-reduction reactions in contaminated environments like groundwater, soil and sediments are often influenced by the physical and chemical properties of the medium, but adding substances like composts and biochar or any other organic and inorganic amendments can help manipulate the oxidation reduction reactions.80, 81 The toxicity as well as mobility of many elements like As, Cr, Hg, Se, Pb, Ni and Cu are also found to be influenced by redox reactions since the chemical reactions are influenced by the oxidation states of these metals.77,82 Some specific microbes that serve as bioremediators for heavy metals are listed in Table 2.
Table (2):
Microorganisms used for bioremediation process.
Microorganisms |
Element/compound |
Citation |
|---|---|---|
Saccharomyces cerevisiae |
Heavy metals, Pb, Hg, Ni |
[85, 86, 87] |
Cunninghamella elegans |
Heavy metals |
[87] |
Pseudomonas fluorescens and Pseudomonas aeruginosa |
Fe2+, Zn2+, Pb2+, Mn2+, Cu2+ |
[88] |
Lysinibacillus sphaericus CBAM5 |
Co, Cu, Cr, Pb |
[89] |
Microbacterium profundi strain Shh49T |
Fe |
[90] |
Aspergillus versicolor, A. fumigatus, Paecilomyces sp., Paecilomyces sp., Terichoderma sp., Microsporum sp., Cladosporium sp. |
Cd |
[91] |
Geobacter sp. |
Fe (III), U (VI) |
[92] |
Bacillus safensis (JX126862) strain (PB-5 and RSA-4) |
Cd |
[93] |
Pseudomonas aeruginosa, Aeromonas sp. |
U, Cu, Ni, Cr |
[94] |
Aerococcus sp., Rhodopseudomonas palustris |
Pb, Cr, Cd |
[95, 96] |
Role of Genetically Engineered Microorganisms (GEMs) in Bioremediation
GEMs are studied to have more degradative capabilities than many natural microbes, in removing persistent compounds in the natural environmental conditions.95-97 With the application of various genetic engineering approaches, the modified genome of the microorganisms have been found to be more efficient in enhancing bioremediation. Methods as single gene editing, pathway construction and change of existing gene sequences aid in modifying rate-limiting stages of the metabolic process.95, 98 GEMs have thus been studied to assist in the removal of heavy metals such as Fe, Cd, As, Cu, Hg and Ni.95, 99-101
GEMs of certain bacteria, fungi and algae have been employed to degrade organic pollutants as oil spills, camphor, hexane, naphthalene, toluene, octane, xylene, halobenzoates, trichloroethylene etc. They are found to be more potent than the natural strains with greater degradative efficiencies and a quick acclimatization to the pollutant medium as substrates.102
Beneficial microorganisms are used widely in various practices. There is a lot of development in technological innovations for safe and environmental friendly disposal of sewage sludge. One such technology referred to as Effective Microorganisms (EM), a brand name developed by Dr. Teruo Higa, a Professor of Horticulture at the University of Ryukus, Okinawa, Japan103, 104 is now being put to use in organic farming, medicine, environment, livestock sector, forestry and agriculture.103, 105 EM is a microbial composition in liquid form consisting of a mix of beneficial and nonpathogenic microbes that coexist between aerobic and anaerobic modes. Currently, investigations are underway for its use in water quality restoration, wastewater treatment, sludge treatment and composting.103, 104
Advantages of Bioremediation
Bioremediation is a universally acceptable biological treatment process for polluted and contaminated environments since the end products are non-toxic and mainly consist of the cell biomass, carbon dioxide and water.106 Complete degradation of a range of pollutants is viable and the products from the process are safe for disposal into the environment.107
Bioremediation is a long-term sustainable, cost-effective, eco-friendly process in comparison to the other technologies employed in the removal of hazardous waste. This can be carried out effortlessly in situ, reducing the threat to the biota and environment in general.106
Disadvantages of Bioremediation
Bioremediation is only applicable to the removal of biodegradable contaminants.107 leaving the non-biodegradable pollutants like plastic persisting in the environment and posing a risk. Biological processes are highly influenced by many factors like the presence of metabolically active microorganisms, suitable environmental factors which can aid microbial growth and an appropriate level of nutrients and contaminants. The survival of microorganisms is based purely on the environmental conditions in which they thrive.108 Extrapolation from the pilot-scale studies to field operations is not ideal in certain conditions thus making it critical to carry out on-site research to initiate bioremediation technologies appropriate for sites with a complex mix of unevenly dispersed pollutants in the environment.108
The technique of biotechnology is environment friendly and cheaper when compared to many pollutant removal mechanisms which are used to transform pollutants into benign substances thereby helping to develop production and disposal processes that are environmentally safe. The application of genetic engineering through exploitation of microorganisms to reduce the toxic load in the environment is paving the way for the future of maintaining the environmental quality thus proving to be a very rewarding option to remediate and manage the polluted environment through microorganisms.
Enhancing the natural biodegradation processes proves to be an effective cost-efficient way of treatment. More potential for advances in this field through more knowledge of how microorganisms respond to different specific environments and assist in degradation will help in reducing the impacts of heavy metal pollution on the environment.
ACKNOWLEDGMENTS
The authors would like to thank Bharathidasan University for providing with all the facilities and resources used for the making of this article.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
Not applicable.
- Kumar R, Chauhan A, Yadav N, Rawat L, Goyal MK. Bioremediation of polluted soil obtained from Tarai Bhavan Region of Uttrakhand, India. J Bioremediat Biodegrad. 2015;6:276.
Crossref - Akahori A, Gabryelak T, Jozwiak Z, Gondko R. Zinc-induced damage to carp (Cyprinus carpio L.) erythrocytes in vitro. Biochem Molec Biol Int. 1999;47(1):89-98.
Crossref - Jinhu L, Liang C, Shuozeng D. Trophic transfer, biomagnification and risk assessments of four common heavy metals in the food web of Laizhou Bay, the Bohai Sea. Sci of The Total Env. 2019;670:508-522.
Crossref - Ali H, Wajid A, Karim U, et al. Bioaccumulation of Cu and Zn in Schizothorax plagiostomus and Mastacembelus armatus from River Swat, River Panjkora and River Barandu in Malakand Division, Pakistan. Pak J Zool. 2017;49(5):1555-1561.
Crossref - Javed M, Usmani N. Accumulation of heavy metals in fishes: a human health concern. Int J Environ Sci. 2011;2(2):659-670.
- Rose M, Fernandes A, Mortimer D, Baskaran C. Contamination of fish in UK fresh water systems: risk assessment for human consumption. Chemosphere. 2015;122:183-189.
Crossref - Islam GMR, Habib MR, Waid JL, et al. Heavy metal contamination of freshwater prawn (Macrobrachium rosenbergii) and prawn feed in Bangladesh: A market- based study to highlight probable health risks. Chemosphere. 2017;170:282-289.
Crossref - Singh A, Sharma RK, Agrawal M, Marshall FM. Health risk assessment of heavy metals via dietary intake of foodstuffs from the wastewater irrigated site of a dry tropical area of India. Food Chem Toxicol. 2010;48(2):611-619.
Crossref - Ali H, Khan Ezzat. Trophic transfer, bioaccumulation, and biomagnification of non-essential hazardous heavy metals and metalloids in food chains/webs – Concepts and implications for wildlife and human health. Human and Ecol Risk Assessment. 2018;25:1353-1376.
Crossref - Dixit R, Wasiullah, Malviya D, et al. Bioremediation of heavy metals from soil and aquatic environment: an overview of principles and criteria of fundamental processes. Sustainability. 2015;7(2):2189-2212.
Crossref - Tchounwou PB, Yedjou CG, Patlolla AK, Sutton DJ. Heavy metals toxicity and the environment. Experientia Supplementum. 2012;101:133-164.
Crossref - Hetzer A, Daughney CJ, Morgan HW. Cadmium ion biosorption by the thermophilic bacteria Geobacillus stearothermophilus and G. thermocatenulatus. Appl and Env Microbiol. 2006;72(6):4020-4027.
Crossref - Renella G, Mench M, Gelsomino A, Landi L, Nannipieri P. Functional activity and microbial community structure in soils amended with bimetallic sludges. Soil Biol Biochem. 2005;37:1498-1506.
Crossref - Rajbanshi A. Study on heavy metal resistant bacteria in Guheswori sewage treatment plant. Our Nature. 2008;6(1):52-57.
Crossref - Pires C, Magan N, Castro PML. Bacterial diversity and heavy metal resistance in a contaminated site in Portugal; 4th European Bioremediation Conference. Chania, Greece, 3-6 September 2008.
https://dspace.lib.cranfield.ac.uk/bitstream/handle/1826/5581/
Carlos_Pires_Thesis_2010.pdf - Igiri BE, Okoduwa SIR, Idoko GO, Akabuogu EP, Adeyi AO, Ejiogu IK. Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: a review. J of Toxicol. 2018;2018:2568038.
Crossref - Bissen M, Frimmel FH. Arsenic-a review. Part I: occurrence, toxicity, speciation, mobility. Acta Hydrochimica et Hydrobiologica. 2003;31(1):9-18.
Crossref - Sankarammal M, Thatheyus A, Ramya D. Bioremoval of Cadmium using Pseudomonas fluorescens. Open J of Water Poll and Treatment. 2014;2:92-100.
Crossref - Cervantes C, Campos-Garcia J, Devars S. Interactions of chromium with microorganisms and plants. FEMS Microbiol Rev. 2001;25(3):335-347.
Crossref - Fashola MO, Ngole-Jeme VM, Babalola OO. Heavy metal pollution from gold mines: Environmental effects and bacterial strategies for resistance. Int J of Env Res Pubic Health. 2016;13(11):1042.
Crossref - Malik A. Metal bioremediation through growing cells. Environ Int. 2004;30(2):261-278.
Crossref - Prabhu S, Poulose EK. Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. Int Nano Lett. 2012;2(1):1-10.
Crossref - Booth SC, Weljie AM, Turner RJ. Metabolomics reveals differences of metal toxicity in cultures of Pseudomonas pseudoalcaligenes KF707 grown on different carbon sources. Front Microbiol. 2015;6:827.
Crossref - Gauthier PT, Norwood WP, Prepas EE, Pyle GG. Metal-PAH mixtures in the aquatic environment: A review of co-toxic mechanisms leading to more-than- additive outcomes. Aq Toxicol. 2014;154:253-269.
Crossref - Olaniran AO, Balgobind A, Pillay B. Bioavailability of heavy metals in soil: Impact on microbial biodegradation of organic compounds and possible improvement strategies. Int J Molec Sci. 2013;14(5):10197-10228.
Crossref - Beveridge TJ, Koval SF. Binding of metals to cell envelopes of Escherichia coli K-12. Appl Env Microbiol. 1981;42(2):325-335.
Crossref - Beveridge TJ, Murray RG. Sites of metal deposition in the cell wall of Bacillus subtilis. J Bacteriol. 1980;141(2):876-887.
Crossref - Beveridge TJ, Fyfe WS. Metal fixation by bacterial cell walls. Can J Earth Sci. 1985;22(12):1893-1898.
Crossref - Beveridge TJ. Role of cellular design in bacterial metal accumulation and mineralization. Ann Rev Microbiol. 1989;43:147-171.
Crossref - Monachese M, Burton JP, Reid G. Bioremediation and tolerance of humans to heavy metals through microbial processes: a potential role for probiotics? Appl Env Microbiol. 2012;8(18):6397-6404.
Crossref - Mueller JG, Chapman PJ, Pritchard PH. Creosote- contaminated sites. Their potential for bioremediation. Env Sci Tech. 1989;23:1197-1201.
Crossref - Lovley DR, Woodward JC, Chapelle FH. Stimulated anoxic biodegradation of aromatic hydrocarbons using Fe (III) ligands. Nature. 1994;370:128-131.
Crossref - Sayler GS, Ripp S. Field applications of genetically engineered microorganisms for bioremediation processes. Curr Opin Biotech. 2000;11(3):286-289.
Crossref - Yang T, Chen ML, Wang JH. Genetic and chemical modification of cells for selective separation and analysis of heavy metals of biological or environmental significance. Trends Anal Chem. 2015;66:90-102.
Crossref - Ayangbenro AS, Babalola OO. A New Strategy for Heavy Metal Polluted Environments: A Review of Microbial Biosorbents. Int J Env Res Public Health. 2017;14(1):94.
Crossref - Fomina M, Gadd GM. Biosorption: current perspectives on concept, definition and application. Biores Tech. 2014;160:3-14.
Crossref - Mosa KA, Saadoun I, Kumar K, Helmy M, Dhankher OP. Potential biotechnological strategies for the cleanup of heavy metals and metalloids. Front Plant Sci. 2016;7:303.
Crossref - Madigan MT, Martinko J, Parker J. Brock Biology of Microorganisms 10th Ed., Upper Sadle River, USA.Pearson Education, Inc., 2003.
- Guine V, Martins JMF, Gaudet JP. Facilitated transport of heavy metals by bacterial colloids in sand columns. J Phys IV France. 2003;107:593-596.
Crossref - Okpokwasili GC. Biotechnology and Clean Environment. Proc. of the 20th Annl. Conf. of the Biotechnology Society of Nigeria (BSN), 14th-17th, November, 2007 at the Ebonyi State University, Abakaliki, Nigeria.
- Tang KHD, Hadibarata T. Microplastics removal through water treatment plants: Its feasibility, efficiency, future prospects and enhancement by proper waste management. Environmental Challenges. 2021;5:100264.
Crossref - Sivaperumal P, Kamala K, Rajaram R. Bioremediation of Industrial Waste Through Enzyme Producing Marine Microorganisms. Adv Food Nutr Res. 2017;80:165-179.
Crossref - Fu Y, Viraraghavan, T. Removal of congo red from an aqueous solution by fungus Aspergillus niger. Adv Environ Res. 2002;7(1):239-247.
Crossref - Rubilar O, Diez MC, Gianfreda L. Transformation of chlorinated phenolic compounds by white rot fungi. Crit Rev Environ Sci Technol. 2008;38(4):227-268.
Crossref - Gianfreda I, Bollag, JM. Isolated enzymes for the transformation and detoxification of organic pollutants. Enzymes in the environment: Activity, ecology and applications, New York: Marcel Dekker. 2002:491-538.
Crossref - Abatenh E, Gizaw B, Tsegaye Z, Wassie M. Application of microorganisms in bioremediation-review. J Environ Microbiol. 2017;1(1):2-9.
Crossref - Bruneel O, Duran R, Casiot C, Elbaz-Poulichet F, Personne JC. Diversity of Microorganisms in Fe-As-Rich Acid Mine Drainage Water of Carnoules, France. Appl Environ Microbiol. 2006;72(1):551-556.
Crossref - Baby V, Ayyasamy PM. Diversity of bacteria associated with the metal resistance from the industrial soil samples of Salem District, Tamil Nadu, India. Int J Curr Res Life Sci. 2018;7(4):1475-1481.
- Valls M, de Lorenzo V. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol Rev. 2002;26(4):327-338.
Crossref - Spain A, Alm E. Implications of microbial heavy metal tolerance in the environment. Reviews in Undergraduate Research. 2013;2:1-6.
- Sarret G, Avoscan L, Carriere M, et al. Chemical forms of selenium in the metal-resistant bacterium Ralstonia metallidurans CH34 exposed to selenite and selenate. Appl Env Microbiol. 2005;71(5):2331-2337.
Crossref - Reith F, Rogers SL, Mc Phail DC, Weeb D. Biomineralization of gold: biofilms on bacterioform. Gold. Sci Tech Appl. 2006;313(5784):233-236.
Crossref - Tsai YP, You SJ, Pai, TY, Chen KW. Effect of cadmium on composition and diversity of bacterial communities in activated sludges. Int J Biodeter Biodegrad. 2005;55(4):285-291.
Crossref - Chen JZ, Tao XC, Xu J, Zhang T, Liu ZL. Biosorption of lead, cadmium and mercury by immobilized Microcystis aeroginosa in a column. Proc Biochem. 2005;40(12):3675-3679.
Crossref - Chen XC, Wang YP, Lin Q, Shi JY, Wu WX, Chen YX. Biosorption of copper(II) and zinc(II) from aqeous solution by Pseudomonas putida CZ1. Colloids Surf B Biointerfaces. 2005;46(2):101-107.
Crossref - Ojuederie OB, Babalola OO. Microbial and Plant- Assisted Bioremediation of Heavy Metal Polluted Environments: A Review. Int J Env Res Public Health. 2017;14(12):1504.
Crossref - Adams GO, Fufeyin PT, Okoro SE, Ehinomen I. Bioremediation, biostimulation and bioaugmention: a review. Int J Environ Bioremed Biodegrad. 2015;3:28- 39.
Crossref - Kumar A, Bisht BS, Joshi VD, Dhewa T. Review on bioremediation of polluted environment: A management tool. Int J Env Sci. 2011;1:1079-1093.
- Madhavi GN, Mohini DD. Review paper on Parameters affecting bioremediation. Int J Life Sci Pharma Res. 2012;2:77-80.
- Abatenh E, Gizaw B, Tsegaye Z, Wassie M. The Role of Microorganisms in Bioremediation- A Review. Open J of Env Biol. 2017;2(1):38-46.
Crossref - Malik ZA, Ahmed S. Degradation of petroleum hydrocarbons by oil field isolated bacterial consortium. Afr J Biotech. 2012;11:650-658.
Crossref - Alwan AH, Fadil SM, Khadair SH, et al. Bioremediation of the water contaminated by waste of hydrocarbon by use of Ceratophyllaceae and Potamogetonaceae plants. J Gen Env Res Conserv. 2013;1:106-110.
- Gomez F, Sartaj M. Optimization of field scale biopiles for bioremediation of petroleum hydrocarbon contaminated soil at low temperature conditions by response surface methodology (RSM). Int Biodeter Biodegrad. 2014;89:103-109.
Crossref - Thapa B, Kumar KC, Ghimire A. A review on Bioremediation of petroleum hydrocarbon contaminants in soil. Kathmandu University. J Sci Engg Tech. 2012;8(1):164-170.
Crossref - Kourkoutas Y, Bekatorou A, Banat IM, Marchant R, Koutinas AA. Immobilization technologies and support materials suitable in alcohol beverages production: A review. Food Microbiol. 2004;21(4):377-397.
Crossref - G uzik U, Hupe r t-Koc ure k K , Kr ysiak M, Wojcieszynska D. Degradation potential of protocatechuate 3,4-dioxygenase from crude extract of Stenotrophomonas maltophilia strain KB2 immobilized in calcium alginate hydrogels and on glyoxyl agarose. BioMed Res Int. 2014;2014:1-8.
Crossref - Guzik U, Hupert-Kocurek K, Marchlewicz A, Wojcieszynska D. Enhancement of biodegradation potential of catechol 1, 2-dioxygenase through its immobilization in calcium alginate gel. Electr J of Biotech. 2014;17(2):83-88.
Crossref - Guzik U, Hupert-Kocurek K, Wojcieszynska D. Immobilization as a strategy for improving enzyme properties-Application to oxidoreductases. Molecules. 2014;19(7):8995-9018.
Crossref - Wojcieszynska D, Hupert-Kocurek K, Jankowska A, Guzik U. Properties of catechol 2,3-dioxygenase from crude extract of Stenotrophomonas maltophilia strain KB2 immobilized in calcium alginate hydrogels. Biochem Engg J. 2012;66:1-7.
Crossref - Dzionek A, Wojcieszynska D, Guzik U. Natural carriers in bioremediation: A review. Electr J of Biotech. 2016;23:28-36.
Crossref - Rivelli V, Franzetti A, Gandolfi I, Cordoni S, Bestetti G. Persistence and degrading activity of free and immobilised allochthonous bacteria during bioremediation of hydrocarbon-contaminated soils. Biodegrad. 2013;24(1):1-11.
Crossref - Bayat Z, Hassanshahian M, Cappello S. Immobilization of microbes for bioremediation of crude oil polluted environments: A mini review. Open Microbiol J. 2015;9:48-54.
- Hudson S, Magner E, Cooney J, Hodnett BK. Methodology for the immobilization of enzymes onto mesoporous materials. J Phys Chem B. 2005;109(41):19496-19506.
Crossref - Lee CA, Tsai YC. Preparation of multiwalled carbon nanotube-chitosan-alcohol dehydrogenase nanobiocomposite for amperometric detection of ethanol. Sensors and Actuators B. 2009;138(2):518- 523.
Crossref - Bleve G, Lezzi C, Chiriatti MA, et al. Selection of non- conventional yeasts and their use in immobilized form for the bioremediation of olive oil mill wastewaters. Biores Tech. 2011;102(2):982-989.
Crossref - Klein S, Avrahami R, Zussman E, Beliavski M, Tarre S, Green M. Encapsulation of Pseudomonas sp. ADP cells in electrospun microtubes for atrazine bioremediation. J Indus Microbiol and Biotech. 2012;39(11):1605-1613.
Crossref - Tandon PK, Singh SB. Redox processes in water remediation. Env Chem Lett. 2016;14(1):15-25.
Crossref - Gadd GM. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiol. 2009;156(3):609-643.
Crossref - Rajapaksha AU, Vithanage M, Ok YS, Oze C. Cr (VI) formation related to Cr (III) -muscovite and birnessite interactions in ultramafic environments. Env Sci Tech. 2013;47(17):9722-9729.
Crossref - Bolan NS, Kunhikrishnan A, Naidu R. Carbon storage in a heavy clay soil landfill site after biosolid application.Sci Total Environ. 2013;465:216-225.
Crossref - Beiyuan J, Awad YM, Beckers F, Tsang DCW, Ok YS, Rinklebe J. Mobility and phytoavailability of As and Pb in a contaminated soil using pine sawdust biochar under systematic change of redox conditions. Chemosphere. 2017;178:110-118.
Crossref - Violante A, Cozzolino V, Perelomov L, Caporale A, Pigna M. Mobility and bioavailability of heavy metals and metalloids in soil environments. J Soil Sci Plant Nutr. 2010;10(3):268-292.
Crossref - Chen C, Wang JL. Characteristics of Zn2+ biosorption by Saccharomyces cerevisiae. Biomed Env Sci. 2007;20(6):478-482.
- Talos K, Pager C, Tonk S, et al. Cadmium biosorption on native Saccharomyces cerevisiae cells in aqueous suspension. Acta Univ Sapientiae Agri Env. 2009;1:20-30.
- Infante JC, De Arco RD, Angulo ME. Removal of lead, mercury and nickel using the yeast Saccharomyces cerevisiae. Revista MVZ Cordoba. 2014;19(2):4141- 4149.
Crossref - Tigini V, Prigione V, Giansanti P, Mangiavillano A, Pannocchia A, Varese GC. Fungal biosorption, an innovative treatment for the decolourisation and detoxification of textile effluents. Water. 2010;2(3):550-565.
Crossref - Paranthaman SR, Karthikeyan B. Bioremediation of heavy metal in paper mill effluent using Pseudomonas spp. J Microbiol Biomed Res. 2015;1:1-5.
- Pena-Montenegro TD, Lozano L, Dussan J. Genome sequence and description of the mosquitocidal and heavy metal tolerant strain Lysinibacillus sphaericus CBAM5. Stds Genomic Sci. 2015;10(2):1-10.
Crossref - Wu YH, Zhou P, Cheng H, et al. Draft genome sequence of Microbacterium profundi Shh49T, an Actinobacterium isolated from deep-sea sediment of a polymetallic nodule environment. Genome Announc. 2015;3(3):1-2.
Crossref - Fazli MM, Soleimani N, Mehrasbi M, Darabian S, Mohammadi J, Ramazani A. Highly cadmium tolerant fungi: Their tolerance and removal potential. J Env Health Sci Engg. 2015;13(19):1-9.
Crossref - Mirlahiji SG, Eisazadeh K. Bioremediation of Uranium via Geobacter spp. J Res Dev. 2014;1(12):52-58.
- Priyalaxmi R, Murugan A, Raja P, Raj KD. Bioremediation of cadmium by Bacillus safensis (JX126862), a marine bacterium isolated from mangrove sediments. Int J Curr Microbiol Appl Sci. 2014;3(12):326-335.
- Sinha SN, Biswas M, Paul D, Rahaman S. Biodegradation potential of bacterial isolates from tannery effluent with special reference to hexavalent chromium. Biotech Bioinf Bioengg. 2011;1(3):381-386.
- Sinha SN, Paul D. Heavy metal tolerance and accumulation by bacterial strains isolated from waste water. J Chem Biol Phys Sci. 2014;4(1):812-817.
- Pande V, Pandey S C, Sati D, Bhatt P, Samant M. Microbial Interventions in Bioremediation of Heavy Metal Contaminants in Agroecosystem. Front Microbiol. 2022;13:824084.
Crossref - de Lorenzo V. Recombinant bacteria for environmental release: what went wrong and what we have learnt from it. Clin Microbiol Infect. 2009;15(1):63-65.
Crossref - Bhatt P, Sethi K, Gangola S, et al. Modeling and simulation of atrazine biodegradation in bacteria and its effect in other living systems. J Biomol Struct Dyn. 2020c;40(7):3285-3295.
Crossref - Diep P, Mahadevan R, Yakunin, AF. Heavy metal removal by bioaccumulation using genetically engineered microorganisms. Front Bioeng Biotechnol. 2018;6:157.
Crossref - D’Souza SF. Microbial biosensors. Biosens Bioelectron. 2001; 16(6): 337-353.
Crossref - Verma N, Singh M. Biosensors for heavy metals. Biometals. 2005;18(2):121-129.
Crossref - Azad MAK, Amin L, Sidik NM. Genetically engineered organisms for bioremediation of pollutants in contaminated sites. Chin Sci Bull. 2014;59(8):703-714.
Crossref - Pant G, Garlapati D, Agrawal U, Prasuna RG, Mathimani T, Pugazhendhi A. Biological approaches practised using genetically engineered microbes for a sustainable environment: A review. J Hazard Mater. 2021;405.
Crossref - Safwat SM, Matta ME. Environmental applications of Effective Microorganisms: a review of current knowledge and recommendations for future directions. J Eng Appl Sci. 2021;68.
Crossref - Talaat NB, Ghoniem AE, Abdelhamid MT, Shawky BT. Effective microorganisms improve growth performance, alter nutrients acquisition and induce compatible solutes accumulation in common bean (Phaseolus vulgaris L.) plants subjected to salinity stress. Plant Growth Regul. 2015;75(1):281-295.
Crossref - Ahn K, Lee KB, Kim YJ, Koo YM. Quantitative analysis of the three main genera in effective microorganisms using qPCR- Korean. J Chem Eng. 2014;31(5):849-854.
Crossref - Sinha SN, Biswas K. Bioremediation of lead from river water through lead-resistant purple-nonsulfur bacteria. Global J Microbiol Biotech. 2014;2(1):11-18.
- Sutar H, Das CK. A Review on: Bioremediation. Int J Res Chem Env. 2012;2(1):13-21.
- Shishir TA, Mahbub N, Kamal NE. Review on Bioremediation: A Tool to Resurrect the Polluted Rivers, Pollution, 5:3 (2019): 555-568.
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.