ISSN: 0973-7510
E-ISSN: 2581-690X
The gradual rise of multidrug resistant micro-organisms is a national concern for all health care providers. Linezolid belongs to the oxazolidinone class of antimicrobials. it is a “last resort” used for the management of gram positive bacterial infections. Developing linezolid resistance creates a great challenge for treating bacterial infections. The objective of the current study is to determine the microbial profile and linezolid resistance in gram positive cocci isolated from blood stream infections. 1855 blood samples were analysed for microbial profile and antimicrobial sensitivity testing in our tertiary care centre over a 6 month period. In using Kirby-Bauer’s disk diffusion method for antimicrobial susceptibility testing, linezolid resistance was detected according to CLSI guidelines. Out of 1855 blood culture samples, 732 (39.4%) were identified to be culture positive. Amongst culture positive isolates mostly (83.3%) gram negative bacteria were isolated, and 16.7% were Gram positive bacterial isolates. Klebsiella species were the most prevalent among gram negative isolates. The linezolid resistance pattern was coagulase negative staph (CONS) was 25%, staphylococcus was 24% and streptococcus was 20%. This study reveals significant linezolid resistance in gram positive bacteria isolated from blood culture. The emergence of linezolid resistance is a major issue for clinicians treating the infection and it will require prompt monitoring of antibiotic policy and antimicrobial stewardship programs.
Blood Stream Infections, Blood Culture, Bacterial Isolates, Linezolid Resistance
Blood stream infections (BSI) are very common in developing countries and it can lead to life threatening complications in critical patients with significant mortality and morbidity, if not diagnosed promptly and treated properly.1 Antimicrobial resistance, especially multidrug MDR in both gram-negative and gram-positive bacteria, rises rapidly leading to the spread of infections and the default to treat them. Antimicrobial resistance was the leading public health issue globally. The average global prevalence rate was 60% or more.2 Despite recent advances in clinical diagnostics, blood culture methods remained the gold standard for the isolation of micro-organism from bacteremia, septicemia and fungemia3 Everninomicins, carbapenems, oxazolidinone, streptogramins and daptomycin were among the newer antibacterial agents used to treat gram positive bacterial infections.4-6
Linezolid is a synthetic antibacterial agent that belongs to the oxazolidinone class, mechanism of action of Linezolid is to inhibit bacterial ribosomal protein synthesis by acting on the translation process and preventing complex formation.7 Linezolid binds to ribosomal RNA (rRNA), especially 23S rRNA of the 50S ribosomal subunit (V-domain) encoded by rDNA genes found in clinically relevant species.8 Linezolid has bacteriostatic activity against staphylococci (MSSA and MRSA), Enterococci (VRE), and pneumococci as well. The minimum inhibitory concentration (MIC) of linezolid is 1-4 mg/ml. It has moderate antimicrobial activity against Moraxella & Bacteroides species (MIC 8 mg/ml) and is also active against anaerobic gram-positive bacilli and cocci, some gram negative anaerobes, Nocardia and Mycobacterium species.9 Cross-resistance between linezolid and other antimicrobials that inhibit protein synthesis is very rare because the binding site of linezolid (23S portion of the 50S subunit) is totally different from the binding sites of others.10
Antimicrobial resistance is continuously increasing day by day. It varies according to regional and geographical location. For the prevention of multidrug resistance, follow strict antimicrobial policy, rational use of antibiotics and conduct various antimicrobial stewardship awareness programmes.
Study Design
This study was conducted in a Gandhi Medical College and associated Hospital, Bhopal.
Samples (blood) were collected from suspected patients with blood infections (bacteremia or septicemia) attending and/or admitted in the tertiary care hospital over a period of 06 months (July 2013 to December 2013).
Samples collection and processing
Blood samples were collected by aseptic precaution and sent to the microbiology laboratory for culture and sensitivity testing. All collected samples for blood culture were included in the study. We have used conventional methods of blood culture (BHI broth) for blood culture testing. Identification of microbial isolates was done by colony characteristics on culture Medias, gram staining examination, and standard biochemical tests. We have performed Catalase test for differentiation of streptococci to staphylococci (streptococci were Catalase negative), also perfumed Slide and tube coagulase test differentiate staphylococcus aureus to CONS.
Antimicrobial susceptibility testing
Kirby-Bauer’s disk diffusion method was used for Antimicrobial susceptibility testing as per Clinical and Laboratory Standards Institute (CLSI) guidelines.11 We have made bacterial suspension comparable with 0.5 McFarland standard was poured on Mueller-Hinton Agar plates and a Linezolid disk (30 µg) was applied. AST plates were incubated at 35–37°C temperature for 24 hours, if the zone of inhibition was 21 mm or less considered the isolates was resistance to linezolid.
Statistical analysis: for statistical analysis we have calculated the confidence interval, percentage and proportion using graph pad software. Calculation of confidence intervals was done by Clopper and Pearson method based on binomial distribution and F distribution. .
Out of a total of 1855 blood samples received, 732 (39.4%) were culture positive, while 1123 (60.6%) were found to be blood culture negative.
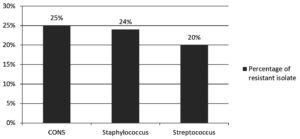
Amongst culture positive isolates, gram negative bacteria accounted for 83.3%, while gram positive bacteria were 16.7%. The predominant bacterial isolates were klebsiella (43.3%) and E.coli (16.5%) among gram negative, while staphylococcus aureus (11.8%) was among gram positive bacterial isolates. A detailed description of bacterial isolates was shown in Table 1. Linezolid resistance was seen in staphylococcus 24%, CONS 20%, and streptococcus 20%. A detailed description of linezolid resistance was shown in Table 2 and Figure.
Table (1):
Frequency and confidence interval of different bacterial isolates of blood culture.
S. No. |
Bacteria isolated |
Number |
Percentage |
95% Confidence Interval |
|---|---|---|---|---|
1 |
Klebsiella |
317 |
43.3% |
0.3968 to 0.4698 |
2 |
E coli |
121 |
16.5% |
0.1391 to 0.1942 |
3 |
Pseudomonas |
76 |
10.3% |
0.0827 to 0.1282 |
4 |
NLFGNB |
54 |
7.3% |
0.0559 to 0.0952 |
5 |
Citrobacter |
36 |
4.9% |
0.0347 to 0.0674 |
6 |
Acinetobacter |
6 |
0.8% |
0.0030 to 0.0178 |
7 |
Staphylococcus |
87 |
11.8% |
0.0963 to 0.1445 |
8 |
CONS |
20 |
2.75 |
0.0168 to 0.0419 |
9 |
Streptococcus |
15 |
2% |
0.0115 to 0.0336 |
Table (2):
Frequency and confidence interval of linezolid resistance among gram positive cocci.
| Resistance pattern to linezolid | |||
|---|---|---|---|
| Bacteria | No. of sensitive isolates | No. of resistant isolates | 95% Confidence interval |
| CONS | 20 | 5 | 0.0866 to 0.4910 |
| Staphylococcus | 87 | 21 | 0.1560 to 0.3450 |
| Streptococcus | 15 | 3 | 0.0433 to 0.4809 |
The rapid emergence of higher and newer antimicrobial resistance is alarming and challenging for people all over the world. In the future era, very few antimicrobial options are available for the treatment of bacterial infections. That’s because the WHO identified emerging antimicrobial resistance as a prime health concern for all over the world.12 To control antimicrobial resistance, precautions such as rational/judicious antibiotic use, prescribing antibiotics after culture-sensitivity reports whenever possible, mounting predicament, and various extensive antibiotic stewardship programmes in developing countries are required.
The current study found a very high blood culture positivity rate (39.5%), which was consistent with Khanal et al.13 and Sharma et al.,14 who reported blood culture positivities of 44% and 33.9%, respectively, but many other studies found very low blood culture positivity, such as Mehdinejad et al.,15 Vanitha et al.,16 and Gohel K et al.
The blood culture positivity rate was higher in the present study. The possible reasons for this were inappropriate use or rational use of antimicrobial agents, emergence of new drug-resistant bacterial strains, antimicrobial administration prior to sample collection and regional variation.
In our study, gram negative bacterial isolates were predominant, accounting for 83.3%, and gram positive isolates were only 16.7%, similar results were also obtained by Paul et al.,18 and Vaghela et al.19 However in contrast to that, other studies where gram positive bacteria were predominantly isolated are Nazir A et al.,20 Tessema B et al.,21 and Pan et al.22
In our study, the most predominant organism isolated from blood culture was Klebsiella followed by E. coli, in concordance to the study conducted by Fahim et al.23
In the current study, gram-positive cocci isolated from blood stream infections were found to have very high linezolid resistance, whereas Mamishi et al.24 found that all isolates were sensitive to linezolid. Higher linezolid resistance in our study could be due to a lower number of isolates, antibiotic overuse, and genetic mutation
In our study, linezolid resistance among coagulase negative staphylococcus (CONS) was 25% (5/20) in accordance with the Panopoulou M et al.,25 observed 20.9% linezolid resistance, Staph epidermidis, in contrast to S. Gandra et al.,26 who found 0.4% resistance.
The current study found linezolid resistance in Staphylococcus aureus was 24% (21/87) A similar finding was also observed by Thool, et al.,27 in contrast to other studies, Comoglu et al.,28 found a higher susceptible rate of linezolid.
Present study observed 20% linezolid resistance streptococci discordance with the Muller-Serieys et al.29 found all streptococcal isolates were sensitive to linezolid.
The emergence of higher linezolid resistance in blood culture isolates is alarming and creates a big challenge for us. A very high prevalence of positive blood culture and linezolid resistance was found in the current study. Antibiotic selection based on culture sensitivity reports, antimicrobial policy development, and various antimicrobial stewardship programmes may all aid in the reduction of multidrug resistance.
Limitation of the Study
CLSI of 2006 was used for antimicrobial susceptibility testing.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SC conceived and designed the study, conducted research, provided research materials, collected and organized data. PS provided logistics support, analyzed and interpreted data. RA wrote the manuscript, corresponds the article. AD reviewed and edited the manuscript.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript
ETHICS STATEMENT
This study was approved by Institutional Ethics Committee, Gandhi Medical College, Bhopal, M.P., India.
- Leibovici L, Samra Z, Konigsberger H, Drucker M, Ashkenazi S, Pitlik SD. Long-term survival following bacteremia or fungemia. JAMA. 1995;274(10):807-812.
Crossref - Oduyebo OO, Olayinka AT, Iregbu KC, et al. A point prevalence survey of antimicrobial prescribing in four Nigerian tertiary hospitals. Ann Trop Pathol. 2017;8(1):42-46.
Crossref - Shanson DC. Blood culture technique: current controversies. J Antimicrob Chemother. 1990;25(Suppl C):17-29.
Crossref - Sader HS, Jones RN, Ballow CH, et al. Antimicrobial susceptibility of quinupristin/dalfopristin tested against Gram-positive cocci from Latin America: Results from the Global SMART (GSMART) Surveillance Study. Braz J Infect Dis. 2001;5(1):21-30.
Crossref - Sader HS, Gales AC. Emerging strategies in infectious diseases: New carbapenem and trinem antimicrobial agents. Drugs. 2001;61(5):553-564.
Crossref - Diekema DJ, Jones RN. Oxazolidinones: A review. Drugs. 2000;59(1):7-16.
Crossref - Shinabarger DL, Marotti KR, Murray RW, et al. Mechanism of action of the oxazolidinones: effects of linezolid and esperezolid on translation reactions. Antimicrob Agents Chemother. 1997;41(10):2132-2136.
Crossref - Matassova NB, Rodnina MV, Endermann R, et al. Ribosomal RNA is the target for oxazolidinones, a novel class of translational inhibitors. RNA. 1999;5(7):939-946.
Crossref - Johnson AP, Warner M, Livermore DM. Activity of linezolid against multi-resistant Gram-positive bacteria from diverse hospitals in the United Kingdom. J Antimicrob Chemother. 2000;45(2):225-230.
Crossref - Lee C-R, Lee JH, Park KS, Jeong BC, Lee SH. Quantitative proteomic view associated with resistance to clinically important antibiotics in Gram-positive bacteria: a systematic review. Front Microbiol. 2015;6:828.
Crossref - Clinical and Laboratory Standards Institute. M7-A7 Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard-seventh edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2006.
- World Health Organization (WHO). Antimicrobial resistance: global report on surveillance. 2014.
- Khanal B, Harish BN, Sethuraman KR, Srinivasan S. Infective endocarditis: Report of a prospective study in an Indian hospital. Trop Doct. 2002;32(2):83-85.
Crossref - Sharma PP, Halder D, Dutta AK, et al. Bacteriological profile of neonatal septicemia. Indian Pediatr. 1987;24(11):1011-1017. PMID: 3450639
- Mehdinejad M, Khosravi AD, Morvaridi A. Study of prevalence and antimicrobial susceptibility pattern of bacteria isolated from blood cultures. J Biol Sci 2009;9(3):249-253.
Crossref - Rani NV, Gopal K, Narendra MV, et al. A retrospective study on blood stream infections and antibiotic susceptibility patterns in a tertiary care teaching hospital. Int J Pharm Pharm Sci. 2012;4(1):543-548.
- Gohel K, Jojera A, Soni S, Gang S, Sabnis R, Desai M. Bacteriological profile and drug resistance patterns of blood culture isolates in a tertiary care nephrourology teaching institute. Biomed Res Int. 2014;2014:153747.
Crossref - Paul M, Bhatia M, Rekha US, Diksha, Omar BJ, Gupta P. Microbiological profile of blood stream infections in febrile neutropenic patients at a tertiary care teaching hospital in Rishikesh, Uttarakhand. J Lab Physicians. 2020;12(2):147-153.
Crossref - Vaghela HG, Duttaroy B, Prajapati KC. Bacteriological profile and antibiogram of blood culture isolates from paediatric patients with special reference to ESBL and MRSA in a tertiary care centre. Indian J Microbiol Res. 2019;6(3):261-265.
Crossref - Nazir A, Sana I, Peerzada BY, Farooq T. Study of prevalence and antimicrobial susceptibility pattern of blood culture isolates from a tertiary care hospital of North India. Int J Res Med Sci. 2018;6(12):4046-4052.
Crossref - Tessema B, Lippmann N, Knüpfer M, Sack U, Konig B. Antibiotic resistance patterns of bacterial isolates from neonatal sepsis patients at University Hospital of Leipzig, Germany. Antibiotics (Basel). 2021;10(3):323.
Crossref - Pan F, Zhao W, Zhang H. Value of time to positivity of blood culture in children with bloodstream infections. Can J Infect Dis Med Microbiol. 2019;2019:5975837.
Crossref - Fahim NAE. Prevalence and antimicrobial susceptibility profile of multidrug-resistant bacteria among intensive care units patients at Ain Shams University Hospitals in Egypt-a retrospective study. J Egypt Public Health Assoc. 2021;96(1):7.
Crossref - Mamishi S, Mohammadian M, Pourakbari B,et al. Antibiotic Resistance And Genotyping Of Gram-Positive Bacteria Causing Hospital-Acquired Infection In Patients Referring To Children’s Medical Center. Infect Drug Resist. 2019;12:3719-3726.
Crossref - Karavassilis V, Zarkotou O, Panopoulou M et al. Wide dissemination of linezolid-resistant Staphylococcus epidermidis in Greece is associated with a linezolid-dependent ST22 clone. J Antimicrob Chemother. 2015;70(6):1625-1629.
Crossref - Gandra S, Mojica N, Klein EY, et al. Trends in antibiotic resistance among major bacterial pathogens isolated from blood cultures tested at a large private laboratory network in India, 2008-2014. Int J Infect Dis. 2016;50:75-82.
Crossref - Thool VU, Bhoosreddy GL, Wadher BJ. Detection of resistance to linezolid in Staphylococcus aureus infecting orthopedic patients. Indian J Pathol Microbiol. 2012;55(3):361-364.
Crossref - Comoglu S, Kaya S, Ceran N, Aksoz S, Ozturk S, Karagoz G. Determination of In Vitro Activity of Linezolid in Resistance Gram-positive Bacteria by E-Test Method. Haydarpasa Numune Med J. 2019;59(1):25-30.
Crossref - Muller-Serieys C, Drugeon HB, Etienne J, et al. Activity of linezolid against Gram-positive cocci isolated in French hospitals as determined by three in-vitro susceptibility testing methods. Clin Microbiol Infect. 2004;10(3):242-246.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.