ISSN: 0973-7510
E-ISSN: 2581-690X
Postprandial hyperglycemia (PPG) is among the earliest signs related to type 2 diabetes. Targeting the α-amylase enzyme responsible for the initial stage of carbohydrate digestion can be an effective strategy to control the PPG. With this objective, about 300 Lactic acid bacteria were obtained from different ethnic fermented foods of Sikkim and screened for α-amylase inhibitor (AAI) activity. Five isolates of Lactobacillus (Lb.) plantarum were found to inhibit α- amylase whose inhibitory potential was similar to that of copper sulfate and acarbose. Maximum AAI activity was observed within 24 hours of incubation. All the isolates were also assessed for other probiotic properties including bacterial adherence to hydrocarbons, utilization of prebiotic components, etc. Among the safety features of the isolates tested, none of the isolates was hemolytic and neither could hydrolyze gelatin. Most of the isolates were sensitive to ampicillin, chloramphenicol, and erythromycin out of twelve antibiotics tested. These isolates also showed good response to in vitro simulation of oro-gastric-intestinal transit which confirms its ability to survive in the gastrointestinal tract. Therefore, these Lb. plantarum isolates can be potential probiotic candidates with a- amylase inhibitory activity which can be further exploited for the management of postprandial hyperglycemia.
Lactobacillus plantarum, dahi, probiotic, alpha-amylase inhibitor
The incidence of Diabetes is increasing each year throughout the world affecting about 46.3 crores people worldwide1. International Diabetes Federation (IDF) has predicted the rise of diabetes by 57.3 crores in 2030 and 70.0 crores by 2045 which is a 51% increase within 25 years2. Diabetes is a multi-factorial physiological condition with many complicated and unknown risk factors. Postprandial hyperglycemia (PPG) is one of the earliest abnormalities associated with type 2 diabetes3. The role of PPG on diabetes has always been neglected by medical practitioners because of its complexity. Studies have indicated that control of PPG is more important than controlling fasting hyperglycemia. An individual destined to develop diabetes remains in a PPG state for 10-12 years before the complete onset of diabetes. Thus controlling PPG at the earlier stage can present a permanent cure for the dreaded disease.
During diabetes development, structural and functional changes in the alimentary canal result in the increased activity of major carbohydrate digestive enzymes, i.e. amylase and glucosidase. Inhibitors of a-amylase (𝛼-1,4-glucan-4-glucanohydrolases), an enzyme responsible for initial stages of carbohydrate digestion, have emerged as the molecules of choice due to their unambiguous and unique mechanism of action on PPG4,5. Moreover, the enzyme inhibitor drugs prescribed to inhibit a-amylase and glucosidase for the treatment of diabetes are not free from side effects6. Therefore, an alternative and complementary method for the management of PPG and diabetes is necessary.
Probiotics are the microorganisms when ingested or used at appropriate cell density imparts a health benefit to the host7. Probiotic microorganisms belonging to genus Bifidobacteria, Lactobacillus8, and Saccharomyces9,10 are the most studied ones. Probiotics have been reported to be helpful in irritable bowel syndrome, inflammatory bowel disease, constipation, antibiotic-associated and acute diarrhoea, hypertension, and diabetes11,12. Therefore, the blend of microbial enzyme inhibitors and probiotic Lactobacillus sp will have twin benefits of the nutritional and pharmacological solution for PPG. As lactobacilli have ‘generally regarded as safe’ (GRAS) status13, enzyme inhibitor producing probiotic organisms would be added feature. So far Lactobacillus isolates from traditional ethnic fermented vegetable and milk products from North-East part of India including Sikkim have not been studied for such property. Hence, the study aims to screen a-amylase inhibitors and other probiotic characteristics from various Lactobacillus isolates from the traditional fermented vegetables and dairy products of Sikkim.
Collection of samples
Altogether thirty samples where five samples each of human stool and milk, two samples each of bamboo shoot, mesu (fermented bamboo), kinema (fermented soybean), gundruk (fermented leafy vegetable of brassica family), cow milk, yak ghew (butter) and eight samples of homemade cow dahi were obtained from different locations of Sikkim. The samples were collected in sterile bags/bottles and were taken to the laboratory in an ice-box.
Isolation of lactic acid bacteria (LAB)
Isolation of LAB was carried out by taking 5 g of sample in a 45 ml sterile saline water (0.85%) and was homogenized in a stomacher (Seward, UK)14. Serial dilutions in the same diluents were prepared and then plated into the Petri-dishes followed by pouring of melted de Man, Rogosa, and Sharpe (MRS) Agar (HiMedia, India) containing 1.5% CaCO3. The inoculated plates were incubated by maintaining the anaerobic condition in Gas Pack System (HiMedia, India) at 37 °C for 72 h. The pure cultures were obtained by streaking on sterile MRS agar plates. The pure cultures of each LAB isolate were preserved -80 °C in MRS broth containing 15 % (v/v) glycerol.
Screening for a-amylase inhibitory (AAI) activity
About 300 LAB isolates were examined for AAI activity as described by Feng et al. with some modifications15. Cell-free supernatant of 18-20 h MRS broth culture of each LAB isolates was obtained by removing the cells by centrifugation. 40 µL of cell-free supernatant was added to 50 µL 0.02 M sodium phosphate buffer (pH 6.9) containing 0.5 mg/ml porcine pancreatic a-amylase (Sigma, US) solution and kept at room temperature for 10 min. 40 µL of 1% starch solution was added to the inhibitor-enzyme mixture and allowed to stand under the same condition. The reactions were then quenched by the addition of 10 µL Lugol’s iodine solution. The intensity of the developed colour was measured in the micro-plate reader (Bio-rad, Switzerland). 40 µL of MRS broth was incubated with the enzyme instead of cell-free supernatant in the control and the blank tube consisted of MRS broth without the enzyme. Acarbose and copper sulfate (1%) was used as a positive control.
The inhibition of a-amylase was calculated as:
Inhibition % = {(Test – Control) / (Blank-Control)} x 100
Production of a-amylase inhibitor
The selected LAB isolates were cultured in 100 ml MRS broth in 250 ml culture flask under shaking condition (60 rpm) at 37 °C. 250 ml of MRS broth in 250 flasks were kept as control under a similar condition. AAI activities of selected LAB isolates were determined by withdrawing the samples at a regular interval of eight hours from 0 to 96 h. The AAI activities were assayed as per the method described in the screening section. Along with AAI activities, pH and optical density of the culture medium and uninoculated MRS broth was also measured at regular intervals.
Stability of crude a –amylase inhibitor
18-20 h MRS broth culture of LAB isolates were centrifuged and the supernatant was collected. The culture supernatant showing AAI activities were stored at room temperature (RT), 8°C and -20°C and AAI activities were checked at regular intervals of seven days to check the stability of AAI active constituent.
Hydrophobicity test
Hydrophobicity was estimated by taking 4 ml of the 24 hr LAB cultures and were centrifuged at 4 °C, 7500 rpm for 5 min. Then the cell pellet was washed twice with Ringer’s solution. Finally, a 9 ml Ringer’s solution was added to resuspend the cells. OD580 was taken by pipetting 1 ml of the cell suspension. Again, an equal volume of the suspension was mixed with an equal volume of the hydrocarbons (chloroform, n-hexadecane, n-octane, p-xylene) and vortexed vigorously. After allowing for 30 min for the separation of the aqueous and the hydrocarbon phases, the aqueous phase was pipetted into a clean cuvette and the OD580 was measured in the spectrophotometer (Lambda-25, Perkin Elmer)16. The hydrophobicity percentage was calculated as
Hydrophobicity % = {(OD before mixing with hydrocarbons – OD after mixing with hydrocarbons)/OD before mixing with hydrocarbons} x 100
Utilization of prebiotics
The utilization of prebiotic components was examined by inoculating the LAB isolates into the MRS broth containing filter-sterilized raffinose, xylitol, inulin, and fructo-oligosaccharides at the level of 2% (w/v) instead of glucose and incubated at 37 °C for 5 days.
Hemolytic activity and gelatin hydrolysis
The overnight grown LAB isolates were streaked on the blood agar plates containing 5% blood and incubated at 37 °C for two weeks. The plates were observed for the formation of hemolytic zones around the LAB colonies17. Gelatin hydrolysis was carried out by inoculating LAB isolates in Brain Heart Infusion (BHI) agar stabs containing 3 % gelatin18. The cultures were kept at 4 °C for half an hour followed by recording the liquid gelatin hydrolysis daily during the incubation period.
Antibiotic susceptibility test
The antibiotic susceptibility was studied by using the method of Bauer et al.19. MRS Broth was inoculated with the LAB isolates and incubated at 37 °C for 24 h. Inoculum density of all the bacterial cultures was maintained as per 0.5 McFarland standard. Staphylococcus aureus and E. coli lawn for reference were made on Mueller Hinton Agar (MHA) while for LAB isolates, LAB susceptibility test medium (LSM) agar plates were used for making bacterial lawn. Using sterile forceps, the antibiotic discs were placed on the agar medium aseptically. Plates were incubated at 37 °C for 24-48 hours under an anaerobic Gas Pack system (HiMedia, India). The zone of inhibitions was measured and was expressed as sensitive, intermediate, and resistant20.
Survival studies during oro-gastric-intestinal transit
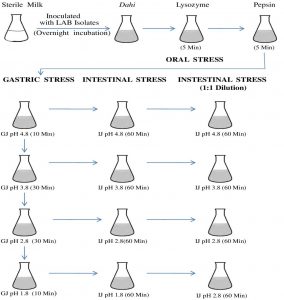
LAB isolates showing the best AAI activities and other probiotic properties were analyzed for their ability to survive the oro-gastric-intestinal transit following the method of Marteau et al21 with some modifications. Actively growing 5 ml overnight culture of LAB isolates was centrifuged at 10,000 rpm and cells were resuspended and washed with 0.85% physiological saline. Finally, 5 ml of cells in physiological saline were inoculated into sterile 500 ml milk and incubated at 37 °C until the milk was coagulated and turned into dahi. The in vitro survival of LAB isolates during oro-gastric-intestinal transit was performed at 37 °C as shown in Fig. 1.
Fig 1. Flow chart showing the plan for oro-gastric-intestinal stress. Time of incubation after each stress is indicated in the parenthesis and all incubations were carried at 37°C. Each pH stress was followed by intestinal stress. GJ, gastric juice; IJ, intestinal juice.
Oral stress was simulated by dilution of the dahi-bacterial mixture with 150 mg/L lysozyme (Sigma-Aldrich) in a sterile electrolyte solution. The gastric environment was mimicked by further dilution and acidification by the addition of gastric juice. pH of the mixture was acidified (addition of 1M HCl) to lower the pH from 4.8 to 3.8, 2.8 and 1.8 and kept in the water bath at 37 °C for 10, 30, 30, and 10 min for each pH value respectively.
Intestinal stress in the small intestine was mimicked by treating 5ml aliquot with intestinal juice for 90 min after and maintaining the pH to 6.5 for 90 min22. Then, 1mL aliquots were withdrawn and analyzed immediately. Subsequently, intestinal juice stressed mixtures were diluted with an equal volume of intestinal electrolyte solution23 reducing the bile to 3g/L. After 90 min, the samples were recovered for analysis. Surviving bacterial populations were enumerated after exposure to the stress conditions.
Identification of lactic acid bacterial isolates
Phenotypic identification of Lactobacillus isolates was done as per Bergy’s Manual of Systematic Bacteriology24. The genotypic identification was done based on 16S rDNA sequences. Phenol chloroform method25 was performed to obtain the genomic DNA from the bacterial isolates. 16S rDNA region was amplified with 27F and 1492R primers. 1 µl of purified PCR products were subjected to sequencing in automated DNA sequencer (3500 Genetic analyzer, Applied Biosystems, Japan). A sequence similarity search was carried out using the BLAST tool from the NCBI website. Phylogenetic analysis of the obtained sequences was performed using UPGMA method26 and the evolutionary distances were analyzed through the Maximum Composite Likelihood method27 using the MEGA7 software28. All the 16S rRNA gene sequences were submitted in GenBank (https://www.ncbi.nlm.nih.gov) and the accession numbers were obtained25.
Analysis of data
At least three independent experiments were conducted to collect the data and the means and standard deviation/error were calculated. Kruskal-Wallis Test was performed to determine the significant differences between means at p < 0.05 followed by Mann–Whitney–Wilcoxon post hoc analysis. All statistical analyses were performed using RStudio.
Screening for AAI activity
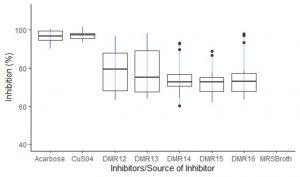
Out of 300 LAB isolates screened for AAI activities, only five isolates from home-made cow dahi were found positive. These isolates showed variable amylase inhibition pattern (Fig. 2) among which the isolate DMR12 and DMR13 had consistent AAI activities while DMR14, DMR15 and DMR16 had some outliers. Though the mean AAI activities of DMR12 (79.2%) were found to be higher than those of DMR13 (78.3%), DMR14 (75.4%), DMR15 (73.1%), and DMR16 (75.5%), statistically, all the AAI activities were similar (p < 0.05). Acarbose (96.8%) and copper sulphate (97.2% ) were taken as positive control shown in Table 1. All the isolates showed excellent AAI activities which were only slightly less and statistically significant (p < 0.05) than those of acarbose and copper sulfate. Similar action on a-amylase and a-glucosidase by the cellular extracts of Lactobacillus sp. isolated from infant faeces and other sources have been reported which were comparable to that of acarbose29,30 The acarbose can show up to 98% inhibition on a-amylase in a dose-dependent manner31,32. This is the reason for its use as one of the prescription drugs for the management of post-prandial hyperglycemia in diabetic patients33. Copper sulfate is also the potent inhibitor of a-amylase enzyme and regularly used as a positive control for the screening of α – amylase inhibitors34.
Fig. 2. Boxplot showing the distribution pattern of AAI activities of the inhibitor from different LAB isolates. AAI activities of MRS broth was below 2% and hence not shown in the plot.
Table (1):
AAI activities of culture-free supernatant of LAB isolates.
Inhibitor |
AAI Activities (%) |
|---|---|
Acarbose |
96.8 ± 2.98a |
Copper sulfate |
97.2 ± 2.31a |
DMR12 |
79.2 ± 11.7b |
DMR13 |
78.3 ± 12.4b |
DMR14 |
75.4 ± 9.58b |
DMR15 |
73.1 ± 7.27b |
DMR16 |
75.5 ± 11.5b |
MRS Broth |
1.97 ± 0.77c |
Data represent the means of five independent replicates.
Different superscripts indicate the means are significantly different (p <0.05)
Production of α -amylase inhibition
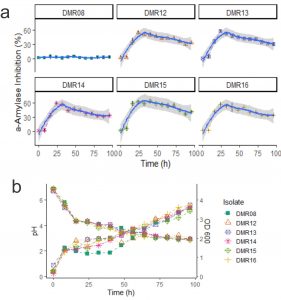
When the amylase inhibitory properties of the isolates were checked during the growth of the AAI positive isolates, DMR15 showed the highest inhibitory property (Fig. 3a). The isolate DMR15 showed 59% of inhibition after 16 hours and reached 62.5% in 24 hours. The isolate DMR08 was considered as negative control which had no AAI activities. All the six LAB isolates including DMR08 decreased the pH of the growth medium as shown in Fig. 3b. The optical density of the growing cells was also similar for all the isolates. AAI activity of all the isolates decreased after 32 hours. The AAI production corresponds to a late exponential phase of the growth for all the isolates. Though DMR08 could not produce a-amylase inhibitor, its growth characteristics and acidification of the medium were similar to those of AAI positive isolates. Moreover, the MRS medium was also negative for amylase inhibitory activity. This indicates that the AAI activity is not because of the acidification of the growth medium or due to the components of the growth medium.
Fig 3. (a) AAI activities during the growth of LAB isolates. Gray ribbon indicates the local regression line calculated using Loess method (b) pH are shown by descending lines and optical density measured during the growth of the isolates are indicated by ascending lines.
Stability of crude a –amylase inhibitor
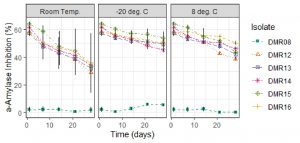
Cell-free culture supernatant showed a decrease in AAI activities at different storage conditions (Fig. 4). AAI activities of crude extract were preserved best at -20°C which remained around 50% after 28 days whereas in 8°C it showed around 45% AAI activities and at room temperature only 35% AAI activities.
Hydrophobicity test
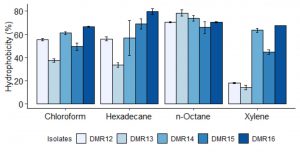
The probiotic bacteria should stay in the GI tract, though for a short period of time35. Therefore, to attach to the intestinal mucosa, the cell surface of the probiotic microorganism should be hydrophobic in nature36,37. All the isolates showed moderate to high ability to adhere to chloroform, n-hexadecane, and n-octane (fig 5). The highest hydrophobicity was shown by DMR16 in n-hexadecane and n-octane which was more than 70%. While in p-xylene, most of the isolates showed high adherence ability except DMR12 and DMR13 (<20%). The adherence to hydrocarbons is dependent on incubation time, the composition of culture media, the presence of organic acids, and on the nature of the solvent used37,38. Partitioning of microbial cells to the hydrocarbon phase indicates the interaction of the cell surface to the mucosa through van der Waals and electrostatic interactions rather than the hydrophobic nature of the cell39.
Fig. 5. Adherence ability of different LAB isolates to hydrocarbons expressed as hydrophobicity (%). Data represent the means of three independent replicates.
Utilization of prebiotics
Prebiotics are nondigestible food components that are utilized by the beneficial colonic bacteria such as Lactobacillus and Bifidobacterium, stimulating their growth and metabolic activity36,40. Inulin consists of D-fructose units linked by a β (2-1) bonds with either a glucose or fructose unit at the end of the chain41. Humans do not have enzymes to hydrolyze the β (2-1) linkages and thus cannot digest it. However, these prebiotic components can only be utilized by certain bacteria present in the colon41, such as by most of the bifidobacteria42 which can produce β-fructosidase enzyme. While the production of β-fructosidase by lactobacilli is rare, nevertheless few isolates of Lb. paracasei, Lb. casei, Lb. acidophilus, and Lb. delbrueckii can produce it43,44. Genomic and proteomic analysis has revealed a functional inulin operon fosRABCDXE in Lb. plantarum responsible for coding β–fructosidase and fructose transport45. In this study, all the five LAB isolates were able to utilize at least three prebiotic components including inulin (Table 2). Moreover, these isolates are also able to utilize low-calorie polyols such as mannitol and sorbitol46. Sorbitol is one of the emerging prebiotic components although its absorption in the upper GI tract is very slow. Thus, a significant proportion of sorbitol can reach the colon to be utilized by the colonic bacteria. In vivo studies in rats have shown that feeding with sorbitol increases the colonic and cecal population of lactobacilli47. The bifunctional alcohol dehydrogenase adhE under the control of transcriptional regulators AcrR and Rex, is responsible for the utilization of mannitol and sorbitol in Lb. plantarum48. A study on the utilization of pectin by probiotic lactobacilli is meager46 although Bifidobacteria have been shown to utilize pectin in a strain-dependent manner49.
Table (2):
Utilization of prebiotic components by LAB isolates having AAI activity.
| Isolate code | Utilization of prebiotics | |||||||
|---|---|---|---|---|---|---|---|---|
| Fruto-oligo -saccharides | Inulin | Mannitol | Raffinose | Sorbitol | Stachyose | Pectin | Xylitol | |
| DMR12 | – | + | + | – | w | – | + | – |
| DMR13 | – | + | – | w | – | + | + | – |
| DMR14 | – | + | + | – | + | – | – | – |
| DMR15 | – | + | + | – | + | – | + | – |
| DMR16 | – | + | + | – | + | + | + | – |
Growth characteristics are represented by w, weakly positive; +, positive; ++, strongly positive; -, no growth
Hemolysis and gelatin hydrolysis
Among the safety aspects, the probiotic microorganisms should neither be hemolytic nor should hydrolyze gelatin. The lactobacilli isolates were screened for hemolysis and gelatin hydrolysis since these are among the important virulence factors present in clinical isolates, and common in Lactobacilli isolates from dairy sources50. In our study none of the LAB isolates was able to cause a- or β-hemolysis and gelatin hydrolysis (data not shown) as reported by Halder et al51.
Antibiotic sensitivity
Probiotic bacteria should also be sensitive to antimicrobial agents or the resistance should not be transferrable to the pathogenic bacteria in the same niche52. Microbial isolates used in the food and feed having transferable antimicrobial resistance are considered as a hazard by the European Food Safety Authority (EFSA)53. All five LAB isolates of homemade dahi were completely sensitive to ampicillin while with erythromycin only DMR12 was resistant (Fig. 6). All the isolates showed intermediate resistance to tetracycline. However, these isolates were completely resistant to amikacin, kanamycin, methicillin, norfloxacin, streptomycin, vancomycin, and several authors have reported that lactobacilli are resistant to most aminoglycosides54. These isolates need further investigation to determine if these resistance genes are transferable or not. Vancomycin resistance is widespread among lactobacilli, though the resistance is not intrinsic55. The presence of multiple drug efflux pumps has been reported in Lactobacillus plantarum and Lactococcus lactis and it is the frequently employed mechanism of resistance56. Aminoglycoside phosphotransferase has been reported from the members of Lb. delbrueckii group and Lb. sakei57,58 which confers resistance in these isolates.
Fig. 6. Antibiotic sensitivity of LAB isolates. Blue dark, purple colour circles indicates the resistant characteristics while Yellow and dark colour circles represent the sensitive characteristics. Conc. of the antibiotics is indicated in parenthesis in the unit of mcg.
Survival studies during oro-gastric-intestinal transit
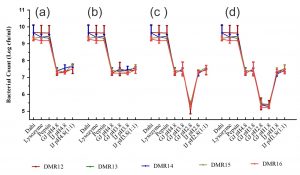
To elicit the full effect in the host, the probiotic candidate microorganism should survive the harsh conditions encountered in the upper part of the digestive tract59,60 Most of the bacteria isolated from food samples are very sensitive to the acidic conditions of the stomach, nevertheless, several members of LAB are acid-tolerant and can grow well in acidic conditions because of the presence of proton extrusion mechanism61. Protection of potential probiotic bacterium by the food matrix i.e. dahi and the effect of lysozyme, the influence of pepsin and the gastric stress, and further the intestinal stress due to the presence of bile, simulating the successive passage of bacteria to the intestine during digestion was performed21. All the isolates showed a very good response to the in vitro simulation of oro-gastric-intestinal transit (Fig. 7). The effect of lysosome and pepsin on bacterial survival was almost nil while with the introduction of gastric juice of pH 4.8, the survival capability decreased by 2 log cycles. There was a good number of surviving populations (5.2 log cfu/ml) even at the lowest gastric pH of 1.8 which was similar to that reported for Lb. acidophilus under similar conditions62. After exposure to gastric juice of pH 1.8, these isolates were subjected to intestinal stress containing pancreatin and bile acids. Nevertheless, these isolates were able to increase its population to 7.5 log cfu/ml as reported by Fernandez et al62,63. Probiotic microorganisms should also tolerate bile salts present in the small intestine and Lactobacillus are resistant to intestinal bile contents to varying degrees64.
Fig. 7. Microbial count (cfu/g) of LAB isolates after exposure to different stress conditions prevailing during oro-gastric intestinal transit. Each figure represents a stress condition up to a particular pH and then exposed to intestinal stress (a) up to pH 4.8 and then exposed to intestinal stress, (b) up to pH 3.8 and then exposed to intestinal stress, (c) up to pH 2.8 and then exposed to intestinal stress, (d) Stress up to pH 1.8 and then exposed to intestinal stress
Bacterial Identification
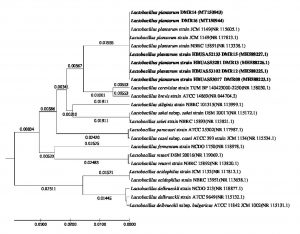
The characterization of the isolates based on morphology and growth characteristics as per the Bergy’s Manual of Systematic Bacteriology24 allowed all the five isolates of LAB to be identified as Lactobacillus sp. (Table 3). Further the species of the bacterial isolates were confirmed by the 16S rRNA gene sequence analysis. The DNA sequences were aligned by the ‘codon code aligner’ and the phylogenetic tree (Fig. 8) was prepared by the UPGMA method using MEGA7 software28. All the LAB isolates were identified as Lactobacillus plantarum upon the comparison of the sequences with the NCBI nr/nt database using the BLAST tool. The sequences were submitted to the GenBank (Table 4).
Fig. 8. Evolutionary relationship of taxa. The evolutionary history was inferred using the UPGMA method1. The optimal tree with the sum of branch length = 0.25083902 is shown. The tree is drawn to scale, with branch lengths (next to the branches) in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method2 and are in the units of the number of base substitutions per site. The analysis involved 25 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 658 positions in the final dataset. Evolutionary analyses were conducted in MEGA73.
Table (3):
Phenotypic characterization of LAB isolates having AAI activities.
| Isolate Code | Growth in/at | Acid production from | Genus | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NaCl (%) | pH | Temperature (⁰C ) | D-Fructose | Galactose | Glycerol | Cellobiose | Maltose | D-Mannitol | Melibiose | Lactose | Rhamnose | D-Ribose | D-Salicin | D-Sorbitol | Sucrose | Trehalose | D-Xylose | |||||||
| 6.5 | 10 | 3.6 | 9.6 | 10 | 15 | 45 | ||||||||||||||||||
| DMR12 | + | – | w | – | – | w | + | + | + | w | + | + | + | w | + | w | + | + | w | + | + | + | Lactobacillus sp | |
| DMR13 | w | – | – | – | + | + | – | + | – | w | w | + | – | – | – | w | + | + | – | + | + | w | Lactobacillus sp | |
| DMR14 | – | – | + | + | – | + | – | + | – | w | + | + | + | + | – | w | + | + | + | + | + | w | Lactobacillus sp | |
| DMR15 | w | – | + | – | – | w | + | + | + | w | + | + | + | w | + | w | + | + | + | + | + | w | Lactobacillus sp | |
| DMR16 | + | – | + | – | – | w | – | + | + | w | w | + | + | + | + | – | + | + | + | + | + | w | Lactobacillus sp | |
Growth characteristics are represented by w, weakly positive; +, positive; ++, strongly positive; -, negative
All the isolates were Gram positive, catalase negative and non-spore forming, homofermentative rods (could not produce gas from glucose)
Except DMR12, none of the isolates produced ammonia from arginine
None of the isolates produced acid from D-arabinose
Table (4):
Bacterial isolates, their 16S identities and NCBI GenBank accession number.
S.No |
Isolate ID |
Bacteria |
Accession No. |
|---|---|---|---|
1 |
DMR12 |
Lactobacillus plantarum |
MH588225.1 |
2 |
DMR13 |
Lactobacillus plantarum |
MH588226.1 |
3 |
DMR14 |
Lactobacillus plantarum |
MT150943 |
4 |
DMR15 |
Lactobacillus plantarum |
MH588227.1 |
5 |
DMR16 |
Lactobacillus plantarum |
MT150944 |
Some of the isolates of Lactobacillus plantarum isolated from homemade cow dahi were able to produce a-amylase inhibitors. This is the first report that describes a-amylase inhibitor production from Lactobacillus isolates of homemade cow dahi. Besides, these isolates also had the potential to adhere to the hydrocarbons indicating their potential to attach to the intestinal epithelium. None of the isolates was hemolytic and neither could hydrolyze gelatin. Lb plantarum also could utilize some of the prebiotic components efficiently than other isolates. Being able to resist the oro-gastric-intestinal environment, Lb plantarum isolates showed the ability to cope with the harsh environment of the GI tract. Therefore, these Lb. plantarum isolates can be the potential probiotic candidate which can be used in the control of PPG and the management of diabetes.
ACKNOWLEDGMENTS
Authors are thankful to Dr. Anil Kumar Puniya, Principal Scientist, National Dairy Research Institute, Karnal, Haryana for providing protocols for the screening of a-amylase inhibitory activity.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors designed the experiments. U. M. performed the experiments. T. D. prepared the manuscript and analysed the data. All authors read and approved the manuscript.
FUNDING
This study was supported by grants of 49.20 lakhs from the Department of Biotechnology, Govt of India (GBT/279/NE/TBP/2011). Rajiv Gandhi National Fellowships-UGC was also awarded to Ranjan Kaushal Tirwa. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and/or the Supplementary Files
- Magliano DJ, Islam RM, Barr ELM, et al. Trends in incidence of total or type 2 diabetes: Systematic review. BMJ. 2019;366:1-12.
Crossref - Saeedi P, Petersohn I, Salpea P, et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract. 2019;157:107843.
Crossref - Maffettone A, Rinaldi M, Fontanella A. Postprandial hyperglycemia: A new frontier in diabetes management? Ital J Med. 2018;12:108-115.
Crossref - Kim YM, Jeong YK, Wang MH, Lee WY, Rhee HI. Inhibitory effect of pine extract on α-glucosidase activity and postprandial hyperglycemia. Nutrition. 2005;21(6):756-761.
Crossref - Toeller M. a-Glucosidase inhibitors in diabetes: efficacy in NIDDM subjects. Eur J Clin Invest. 1994;24:31-35.
Crossref - Evans JL, Rushakoff RJ. Oral pharmacological agents for type 2 diabetes: sulfonylureas, meglitinides, metformin, thiazolidinediones, α-glucosidase inhibitors, and emerging approaches.
- Hill C, Guarner F, Reid G, et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506-514.
Crossref - Gueimonde M, Salminen S. New methods for selecting and evaluating probiotics. Dig Liver Dis. 2006;38(SUPPL. 2):242-247.
Crossref - van der Aa Kuhle A, Skovgaard K, Jespersen L. In vitro screening of probiotic properties of Saccharomyces cerevisiae var. boulardii and food-borne Saccharomyces cerevisiae strains. Int J Food Microbiol. 2005;101(1):29-39.
Crossref - Sourabh A, Kanwar SS, Sharma OP. Screening of indigenous yeast isolates obtained from traditional fermented foods of Western Himalayas for probiotic attributes. J Yeast Fungal Res. 2011;2(8):117-126.
- Khalesi S, Bellissimo N, Vandelanotte C, Williams S, Stanley D, Irwin C. A review of probiotic supplementation in healthy adults: helpful or hype? Eur J Clin Nutr. 2019;73(1):24-37.
Crossref - Weichselbaum E. Probiotics and health: A review of the evidence. Nutr Bull. 2009;34(4):340-373.
Crossref - Wessels S, Axelsson L, Bech Hansen E, et al. The lactic acid bacteria, the food chain, and their regulation. Trends Food Sci Technol. 2004;15(10):498-505.
Crossref - Tamang JP, Tamang B, Schillinger U, Franz CMAP, Gores M, Holzapfel WH. Identification of predominant lactic acid bacteria isolated from traditionally fermented vegetable products of the Eastern Himalayas. Int J Food Microbiol. 2005;105:347-356.
Crossref - Feng GH, Richardson M, Chen M shun, Kramer KJ, Morgan TD, Reeck GR. a-Amylase Inhibitors from Wheat : Amino Acid Sequences and Patterns of Inhibition of Insect and Human a-Amylases. Insect Biochem Molec BioL. 1996;26(5):419-426.
Crossref - Rosenberg M, Gutnick D, Rosenberg E. Adherence of bacteria to hydrocarbons: A simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett. 1980;9:29-33.
- Isenberg H. Hemolysis on sheep blood agar. In: Clinical Microbiology Procedures Handbook. Vol. 1. Washington D.C: American Society of Microbiology. 1992:1.20.16-1.20.17.
- Isenberg H. Gelatin liquefaction test. In: Clinical Microbiology Procedures Handbook, Vol. 1. Washington, DC.: American Society of Microbiology; 1992:1.19.42-1.19.43.
- Bauer AW, Kirby WMM, Sherris JC, Turck AM, Graevenitz A Von. 40 Microbiology: A Centenary Perspective 1966 Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am J Clin Pathol. 1978;45(3):493-496.
- Charteris WP, Kelly PM, Morelli L, Collins JK. Antibiotic susceptibility of potentially probiotic Bifidobacterium isolates from the human gastrointestinal tract. Lett Appl Microbiol. 1998;26(5):333-337.
Crossref - Marteau P, Minekus M, Havenaar R. Survival of Lactic Acid Bacteria in a Dynamic Model of the Stomach and Small Intestine : J Dairy Sci. 1997;80(6):1031-1037.
Crossref - Fernandes ML, Perin LM, Todorov SD, Nero LA, De Alencar ER, De Aguiar Ferreira M. In vitro evaluation of the safety and probiotic and technological potential of pediococcus pentosaceus isolated from sheep milk. Semin Agrar. 2018;39(1):113-131.
Crossref - Marteau P, Minekus M, Havenaar R, Huis in’t Veld JH. Survival of lactic acid bacteria in a dynamic model of the stomach and small intestine: validation and the effects of bile. J Dairy Sci. 1997;80(6):1031-1037.
Crossref - Vos P De, Garrity GM, Jones D, et al. Bergy’s Manual of Systematic Bacteriology. 2nd Vol. 3. (Whitman WB, Parte AC, Goodfellow M, et al., eds.). Dordrecht: Springer; 2009.
- Green M, Sambrook J. Molecular Cloning A Laboratory Manual. Vol 1. Forth. (Mamiatis T, Fritsch EF, Sambrook J EJ, ed.). New York: Cold Spring Harbor Laboratory Press; 2012.
- Sneath P, Sokal R. Numerical Taxonomy. San Francisco: Freeman; 1973.
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci. 2004;101:11030-11035.
Crossref - Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870-1874.
Crossref - Ramchandran L, Shah NP. Proteolytic profiles and angiotensin-I converting enzyme and α-glucosidase inhibitory activities of selected lactic acid bacteria. J Food Sci. 2008;73(2):M75-M81.
Crossref - Panwar H, Calderwood D, Grant IR, Grover S, Green BD. Lactobacillus strains isolated from infant faeces possess potent inhibitory activity against intestinal alpha- and beta-glucosidases suggesting anti-diabetic potential. Eur J Nutr. 2014;53(7):1465-1474.
Crossref - Mohamed EAH, Siddiqui MJA, Ang LF, et al. Potent a-glucosidase and a-amylase inhibitory activities of standardized 50% ethanolic extracts and sinensetin from Orthosiphon stamineus Benth as anti-diabetic mechanism. BMC Complement Altern Med. 2012;12(1):176.
Crossref - Li K, Yao F, Xue Q, et al. Inhibitory effects against α-glucosidase and a-amylase of the flavonoids-rich extract from Scutellaria baicalensis shoots and interpretation of structure–activity relationship of its eight flavonoids by a refined assign-score method. Chem Cent J. 2018;12(1):82.
Crossref - DiNicolantonio JJ, Bhutani J, O’Keefe JH. Acarbose: safe and effective for lowering postprandial hyperglycaemia and improving cardiovascular outcomes. Open Heart. 2015;2(1):e000327.
Crossref - Safarik I. Rapid Detection of Alpha-Amylase. J Enzym Inhib. 1990;3:245-247.
Crossref - Iniguez-Palomares C, Perez-Morales R, Acedo-Felix E. Evaluation of probiotic properties in Lactobacillus isolated from small intestine of piglets. Rev Latinoam Microbiol. 2008;49(3-4):46-54.
- El-Nagar G, Clowes G, Tudorica C, Kuri V, Brennan C. Rheological quality and stability of yog-ice cream with added inulin. Int J Dairy Technol. 2002;55:89-93.
Crossref - Shakirova L, Grube M, Gavare M, Auzina L, Zikmanis P. Bifidobacterium lacis Bb12 cell surface hydrophobicity and survival of the cells under adverse environmental conditions. J Nutr. 2013;40:85-93.
- Wu. Q, Shah NP. Effects of elaidic acid, a predominant industrial trans fatty acid, on bacterial growth and cell surface hydrophobicity of lactobacilli. J Food Sci. 2014;79:2485–2490.
Crossref - Martienssen M, Reichel O, Kohlweyer U. Surface properties of bacteria from different wastewater treatment plants. Acta Biotechnol. 2001;21:207-225.
- Forssten SD, Sindelar CW, Ouwehand AC. Probiotics from an industrial perspective. Anaerobe. 2011;17(6):410-413.
Crossref - Roberfroid MB, Delzenne NM. Dietary Fructans. Annu Rev Nutr. 1998;18(1):117-143.
Crossref - Roberfroid MB, Van Loo JAE, Gibson GR. The Bifidogenic Nature of Chicory Inulin and Its Hydrolysis Products. J Nutr. 1998;128(1):11-19.
Crossref - Tsujikawa Y, Nomoto R, Osawa R. Difference in Degradation Patterns on Inulin-type Fructans among Strains of Lactobacillus delbrueckii and Lactobacillus paracasei. Biosci Microbiota, Food Heal. 2013;32(4):157-165.
Crossref - Velikova P V, Gotcheva VG, Petrova PM. Novel BulgarianiLactobacillus strains ferment prebiotic carbohydrates. In: J. BioSci. Biotech.; 2014:55-60.
- Buntin N, Hongpattarakere T, Ritari J, et al. An Inducible Operon Is Involved in Inulin Utilization in Lactobacillus plantarum Strains, as Revealed by Comparative Proteogenomics and Metabolic Profiling. Appl Environ Microbiol. 2017;83(2).
Crossref - Yeo SK, Liong MT. Effect of prebiotics on viability and growth characteristics of probiotics in soymilk. J Sci Food Agric. 2010;90(2):267-275.
Crossref - Sarmiento-Rubiano LA, Zuniga M, Perez-Martinez G, Yebra MJ. Dietary supplementation with sorbitol results in selective enrichment of lactobacilli in rat intestine. Res Microbiol. 2007;158(8-9):694-701.
Crossref - Yang X, Teng K, Su R, et al. AcrR and Rex Control Mannitol and Sorbitol Utilization through Their Cross-Regulation of Aldehyde-Alcohol Dehydrogenase (AdhE) in Lactobacillus plantarum. Appl Environ Microbiol. 2019;85(4). doi:10.1128/AEM.02035-18″
- Slovakova L, Duskova D, Marounek M. Fermentation of pectin and glucose, and activity of pectin-degrading enzymes in the rabbit caecal bacterium Bifidobacterium pseudolongum. Lett Appl Microbiol. 2002;35:126-130.
- Kaktcham NF, Zambou FM, El-Soda TM. Antimicrobial and safety properties of lactobacilli isolated from two Cameroonian traditional fermented foods. Sci Pharm. 2012;80:189-204.
Crossref - Halder D, Mandal M, Chatterjee SS, Pal NK, Mandal S. Indigenous probiotic Lactobacillus isolates presenting antibiotic like activity against human pathogenic bacteria. Biomedicines. 2017;5(2):1-11.
Crossref - Jose NM, Bunt CR, Hussain MA. Implications of Antibiotic Resistance in Probiotics. Food Rev Int. 2015;31(1):52-62.
Crossref - EFSA/EDC. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018;16(2):5182.
- Rozman V, Mohar Lorbeg P, Accetto T, Bogovic Matijasic B. Characterization of antimicrobial resistance in lactobacilli and bifidobacteria used as probiotics or starter cultures based on integration of phenotypic and in silico data. Int J Food Microbiol. 2020;314(October 2019):108388.
Crossref - Holliman RE, Bone GP. Vancomycin resistance of clinical isolates of lactobacilli. J Infect. 1988;16(3):279-283.
Crossref - Wacher-Rodarte M del C, Trejo-Munuzuri TP, Montiel-Aguirre JF, et al. Antibiotic resistance and multidrug-resistant efflux pumps expression in lactic acid bacteria isolated from pozol, a nonalcoholic Mayan maize fermented beverage. Food Sci Nutr. 2016;4(3):423-430.
Crossref - Campedelli I, Mathur H, Salvetti E, et al. crossm Genus-Wide Assessment of Antibiotic Resistance in Lactobacillus spp. p. Appl Environ Microbiol. 2019;85(1):e01738-18.
Crossref - Rodriguez C, Petrelli L, Ramirez MS, Centron D, Hebert EM, Saavedra L. Genomic overview of acquired antibiotic resistance mechanisms in Lactobacillus. In: Ruzal SM, ed. Lactobacillus Genomics and Metabolic Engineering. Buenos Aires, Argentina: Caister Academic Press; 2019:185-204.
Crossref - Afzaal M, Saeed F, Saeed M, et al. Survival and stability of free and encapsulated probiotic bacteria under simulated gastrointestinal conditions and in pasteurized grape juice. J Food Process Preserv. 2019;(July):1-8.
Crossref - Havenaar R, Brink B Ten, Huis In ’t Veld JHJ. Selection of strains for probiotic use. In: Probiotics: The Scientific Basis. Dordrecht: Springer Netherlands; 1992:209-224.
Crossref - Conway P, Gorbach S, Goldin B. Survival of Lactic Acid Bacteria in the Human Stomach and Adhesion to Intestinal Cells. J Dairy Sci. 1987;70(1):1-12.
Crossref - Fernandez MF, Boris S, Barbes C. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J Appl Microbiol. 2003;94(3):449-455.
Crossref - Rolim FRL, dos Santos KMO, de Barcelos SC, et al. Survival of Lactobacillus rhamnosus EM1107 in simulated gastrointestinal conditions and its inhibitory effect against pathogenic bacteria in semi-hard goat cheese. LWT – Food Sci Technol. 2015;63(2):807-813.
Crossref - Tokatl M, Gulgor G, Bagder Elmac S, Arslankoz Isleyen N, Ozcelik F. In vitro Properties of Potential Probiotic Indigenous Lactic Acid Bacteria Originating from Traditional Pickles. Biomed Res Int. 2015.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.