ISSN: 0973-7510
E-ISSN: 2581-690X
Scouting for novel and plant-derived biomolecules from endophytic microbial sources draws greater focus on the discovery of novel bioactive metabolites. With this rationale, we scouted the endophytic fungi for taxol, an anticancer diterpenoid and fluorescent biomolecules. In the present study, about 31 endophytic fungal isolates recovered from the Mangifera indica leaves were screened for taxol production in M1D medium. About five isolates were shortlisted based on the thin layer chromatographic analysis of the fungal extracts. Among them Colletotrichum sp. MIP-5 has been identified as a producer of fungal taxol based on UV, FTIR, TLC and HPLC analysis. The partially purified fungal taxol showed similar spectral and chromatographic features of commercially available paclitaxel. In addition to this, we also report the production of a fluorescent compound by Penicillium sp. MIP-3. The Flavin-like compound exhibited a bright greenish-yellow fluorescence with an emission maximum in the range of 505 – 545nm. GC-MS analysis showed the occurrence of Latia luciferin, primarily associated with the bioluminescence of freshwater limpet Latia neritoides. This is the first report of this compound from Penicillium sp. In addition, therapeutically active steroid (β-Sitosterol, Stigmasterol, Campesterol), quinones (Benzo[h]quinoline, 2,4-dimethyl-) and phloroglucinol (Aspidinol) derivatives were also identified from Penicillium sp. MIP-3 based on GC-MS analysis. These molecules could potentially be used in biological and pharmaceutical applications in future.
Bioprospecting, Taxol, Luciferin, Fluorescence, anticancer
Endophytic fungi, an enormously diverse group, habituating the living internal tissues of plants without causing any immediate overt negative effects, represent a rich source of novel natural products.1 Interactions of these cryptic organisms with their host plants vary coherently from mutualism to parasitism depending on the refined balance between the demands of the invaders and the host response.2 In addition, the multitude of interactions among endophytic fungal partners and other biotic components of the niche yields a repertoire of diverse secondary metabolites.3 The biological activity of these metabolites range from biosynthesis of defence compounds (antimicrobial, insecticidal, etc.,) which can exert activity directly or trigger (precursors) /modulate (epigenetic modulators) the hosts defence pathways to confer host benefits (fitness and protection).4 Also, they were documented to modulate the host microbiome through quorum sensing5,6 and confer stress mitigation through host adapted endophytic fungal symbiosis.4 Considerable interest on bioprospecting them for wider applications such as agricultural, industrial and pharmaceutical has been well documented.7-9
Endophytic fungi represent a treasure trove of diverse metabolites and earns special attention as a source of novel bioactive compounds of immense therapeutic applications.10-12 They were also reported to produce plant mimetic metabolites and novel structural metabolites.13,14 Discovery of the diterpenoid anticancer drug paclitaxel (Taxol), from an endophytic fungus (Taxomyces andreanae) of Pacific Yew (Taxus brevifolia)15 had drawn greater attentions to the hidden endophytic fungi as a potential source of therapeutic biomolecules.9,16-18 Elaborate reviews summarizing the chemical (alkaloids, flavonoids, fatty acids, sterols, glycosides, poly heterocyclic compounds, small peptide conjugates, etc.),19-22 and functional (antioxidant, anticancer, antimicrobial, antiviral, etc.),22-26 diversities of the endophytic fungal metabolites have enriched the knowledge about these novelties and aids in developing new drugs based on their pharmacophores.3,27 Studies on bioluminescent28,29 and fluorescent30-32 fungal metabolites were relatively few and unravelling them offers a potential scope for furthering the real-time in vivo monitoring of biological functions in living systems.33-35
Fungi not only manifest diverse metabolic profile, but also exhibit pleomorphic and phenotypic plasticity in nature.36,37 Owing to these phenomenon and subjectivity associated with the description of morphological features employed in the identification processes, molecular identification methods were widely embraced to provide much more robust solution for taxonomic queries. ITS-based molecular identification continue to be in practice for about three decades and it’s been reviewed in length.38,39 The vastness of the fungal ITS data in public domains encompassing most of the documented taxa also renders an excellent opportunity to obtain a well-curated reference dataset against which query sequences could be compared.39
In this study, endophytic fungal flora from Mangifera indica, a perennial horticultural tree with excellent nutritive and medicinal values, was subjected to investigation in this study. Earlier reports have suggested the presence of diverse bioactive metabolites82 and vibrant endophytic fungal community83 in the M. indica. Investigations on the bioactive metabolites of these fungal communities were minimalistic and none could be attributed to taxol related metabolites, hence the endophytic fungi recovered from mango leaves were screened for production of taxol, a diterpenoid anticancer metabolite. The Serendipitous observation of fluorescence in the culture filtrate of one of our fungal isolates motivated to explore them further. The potential taxol producers and fluorescence emitting isolates were identified based on phylogenetic analysis of ITS sequences.
Plant Collection and Surface sterilization
Healthy (Asymptotic) and mature leaves of Mangifera indica were collected from Madurai Kamaraj University campus; in sterile polythene bags and transported carefully to the laboratory.
The leaf samples were subjected to a surface sterilization procedure which involves initial wash in running tap water to remove soil particles and adhered debris, and subsequently washed with distilled water. This was followed by washing in 70% ethanol for 30 seconds, later soaked in 4% sodium hypochlorite for 90 seconds and finally, the leaves were rinsed thrice in sterile distilled water.40
Thoroughly surface sterilized leaves were chopped into small uniformed squares using a sterile scalpel. The leaf segments were inoculated into the PDA media under aseptic condition and incubated in growth chamber. After 72hrs of incubation in 12hr/12hr:light/dark cycle at 25°C, fungal hyphae emerging from leaf segments were transferred to new PDA plates for establishing their axenic culture. A control PDA plate with imprints of surface sterilized leaf segments were maintained all along the study to test the efficacy of surface sterilization protocol.40
Secondary metabolites production
Seven days old fungal biomass was cut from the agar plate in to 1cm2 of area and was aseptically transferred to the 100ml of sterile M1D media prepared in 250ml of conical flask for secondary metabolite production. The inoculated flasks were incubated in static condition for 21 days at 25˚C (± 2˚C) with 12 h light/12 h dark cycle. After 21 days of incubation, the fungal culture fluid was filtered through three layers of filter paper to remove mycelia. The extra cellular fungal compounds in the culture filtrate were extracted with organic solvent Dichloromethane (DCM) in ratio 1:2 V/V (Fungal extract: DCM respectively).41 The organic phase was collected and the solvent was then removed by evaporation under reduced pressure using rotary vacuum evaporator. A portion of dry solid residue was re-dissolved for bioprospecting studies.
Screening and Characterization of taxol
Thin Layer Chromatography
The fungal metabolites condensates recovered in the previous step was analysed on 1mm silica coated TLC plate (Stationary Phase). TLC jar was properly equilibrated with the solvent system of Chloroform: Methanol (9.2:0.8 V/V) (Mobile Phase), as reported by Chakravarthi et al.18 for taxol screening. Analytes were carefully spotted as small uniform spot along with the authentic paclitaxel (Sigma Aldrich) on TLC plate and co-developed in the above mentioned solvent system. TLC plate was taken out from the tank, and visualized in UV illuminator. Putative taxol and paclitaxel would fluoresce blue under UV. Corresponding bands were scrapped and eluted with methanol for further analysis. The retention factor (Rf) was calculated with the formula, Rf = Distance travelled by analyte / Distance travelled by solvent front.
Spectral and Chromatographic analysis
Putative taxol samples resuspended in methanol were analysed using UV-Visible spectrophotometry (HITACHI U-2900 spectrophotometer) and Fourier Transform Infra-Red spectroscopy (Shimadzu FT IR 8000). The spectrums were compared with the standard paclitaxel to ascertain the identity. Also the partially purified putative taxol was analysed by high performance liquid chromatography (Shimadzu shim-pack CLC ODS (4.6 mm x 15 mm), liquid pump-LC-6AD, system controller-SCL-6B, UV–Vis detector (195–700 nm)-SPD6AV, data processor-CR-5A) using mobile phase consisting of methanol:water (65:35) at flow rate of 1 ml/min. Injected sample (20 µl) were analysed at 227nm and the retention time of the recorded peaks were compared against the paclitaxel.42,43
Screening and Characterization of Fluorescent compound
Thin Layer Chromatography
The DCM extracts of three fungal isolates which showed fluorescent bands under UV illumination were re-developed on TLC plate with Chloroform: Methanol (8:2 V/V) (Mobile Phase). The most prominent fluorescent band was scrapped out and eluted with methanol. The analyte was subjected to slow evaporation and subjected to further spectral and chromatographic analysis.
Spectrofluorometric analysis
The partially purified compound was re-dissolved in DCM and subjected to spectrofluorophotometric analysis (Cary Eclipse Fluorescence spectrometer) in synchronous mode (300nm – 700nm) to determine the excitation and emission maxima of the compound.44
Solvatochromism
The partially purified compound (10mg) was dissolved separately in eleven solvents (Hexane, Toluene, Dichloromethane, Chloroform, Ethyl acetate, Acetone, Methanol, Acetonitrile, Acetic acid, Dimethyl sulfoxide and water) differing in their polarity index. After gentle mixing, the mixtures were observed under UV illuminator and fluorescence intensities were monitored using spectrofluorometry.45
GC-MS analysis
The DCM extract of the fungi (MIP-3) with prominent fluorescence was subjected to GC-MS analysis (Agilent GC 7890A / MS5975C) using Agilent DB5MS column of 30m (length) x 0.25mm (internal diameter) x 0.5µm (film thickness) and Helium gas (99.99% pure) at a flow rate of 1 ml/min was used as the carrier gas. For Mass detection, an electron ionization energy method was employed operating at 70 eV (electron Volts) with 0.2 s of scan time and fragments ranging from 40 to 600 m/z. The injection quantity of 1 μL was used (split ratio 5:1), and the injector temperature was maintained at 250°C (constant). The column oven temperature was set at 50°C for 1 min, raised at 12°C per min and final temperature was increased to 300°C for 2 min. The mass fingerprints obtained were matched with the spectral database from the National Institute of Standards and Technology (NIST) library.46
Identification of fungal isolates
The two fungal isolates MIP-3 (producing fluorescent compound) and MIP-5 (potential taxol producer) were subjected to morphological (spore and culture features) and ITS based molecular identification. The mycelial and spore characteristics were recorded under light microscopy (Olympus CX23). Genomic DNA of axenic cultures was extracted using the method described by Cenis47 and was utilized for PCR amplification of ITS region. Universal ITS primers [(ITS1 5’TCCGTAGGTGAACCTGCGG 3’) and (ITS4 5’TCCTCCGCTTATTGATATGC 3’)]48 were used to amplify the internal spacer region spanning the ITS1-5.8S-ITS2 segment. A 25μl reaction mixture comprised of 20ng genomic DNA, 12.5 μl of 2x DNA master mix (Amplicon), 1μl each of the forward and reverse primers (Sigma Aldrich). PCR amplification was performed in Biorad T100 instrument according to the following reaction conditions: 4 min at 94°C for initial denaturation, 30 cycles each of 30 seconds at 94°C for denaturation, 1min at 58.2°C for annealing, 2 min at 72°C for extension, and a final extension at 72°C for 7min. The amplicons were verified on 1.5% agarose gels, purified and sequenced using the same primers at Eurofins Pvt. Ltd, Bangalore, India. Sequences from both the reads were merged to get the full-length sequences as detailed earlier.49 They were blast searched among sequences of type specimens available in NCBI database to identify the closest similar sequences. The closely related sequences were chosen for phylogenetic reconstruction through the maximum likelihood method, with 1000 bootstrap iteration in Mega X. Also, the nucleotide substitution model (Kimura2 + G) best defining the dataset was chosen using the inbuilt options to construct the phylogenetic tree. The generated phylogenetic tree was redrawn using iTOL v6 (https://itol.embl.de/).
Thirty-one endophytic fungal isolates were recovered from the Mangiferae indica leaves and were presented in Fig. 1. Axenic cultures were stored and maintained in the lab repository for future studies.
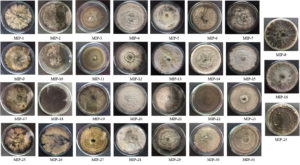
Fig. 1. The culture morphology of 31 endophytic fungal isolates recovered from Mangifera indica leaves.
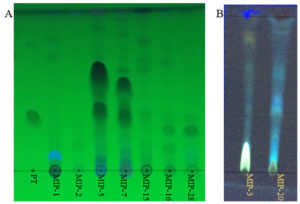
Fig. 2. Thin layer chromatograms (A) shows the DCM extracts of fungal isolates with putative taxol bands. PT denotes the paclitaxel (Sigma-Aldrich). MIP denotes the fungal extracts from different isolates. (B) shows the fluorescent bands in the DCM extracts of fungal isolates (MIP-3 and MIP-20)
Screening and Characterization of Putative Taxol
Of the 31 isolates grown in M1D medium, on TLC based screening, bands of 5 isolates co-migrated with the authentic paclitaxel at same Rf (0.40) and were considered to be potential producers of taxanes (Fig. 2). Among the five isolates, MIP-5 isolate exhibited spectral and characteristic features of authentic paclitaxel (Fig. 3). It had absorption maxima at 237nm and 275nm in UV-visible spectroscopy. In FTIR spectrum, a broad peak at 3375-3390 cm-1 showed the presence of OH stretching in the compound, while a peak at 2945 cm-1 revealed the presence of CH stretching and peaks at 2364 and 1454 cm-1 depicts the NH stretching. The COO stretching peaks were observed at 1383 and 671 cm-1 while peaks in the 1114-1028 cm-1 were predominantly due to the presence of aromatic C and H bends. These peaks correspond to the fingerprints of authentic paclitaxel. Also the HPLC data recorded for the putative taxol from MIP-5 isolates had a retention time around 4.4 min and it was in consistence with the data recorded for paclitaxel.
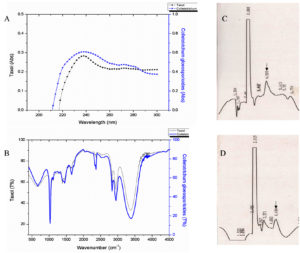
Fig. 3. Shows the data of spectral and chromatographic analysis of putative fungal taxol from Colletotrichum sp. MIP-5 and Paclitaxel. (A) UV-Visible spectrum (B) FTIR spectrum and (C & D) HPLC Chromatogram of Paclitaxel and putative fungal taxol from Colletotrichum sp. MIP-5.
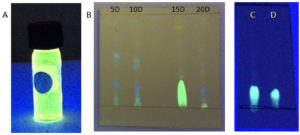
Fig. 4. (A) DCM extract of Penicillium sp. MIP-3 showing fluorescence. (B) TLC analysis of extracts harvested at different days (5th, 10th 15th & 20th day). (C) partially purified fraction resolved on pre-coated silica plates, (D) authentic riboflavin.
Screening and Characterization of Fluorescent molecules
The DCM extract of MIP-3 isolate fluoresced under UV illumination (Fig. 2) and hence was subjected to further analysis. Maximum production of the fluorescent molecule was observed to be during the 15th day of growth in M1D medium (Fig. 4). The fluorescent band observed on the TLC plate was scrapped and eluted in methanol. The partially purified fluorescent molecule dissolved in DCM exhibited an excitation maximum at 400nm and emission in the range of 518nm (Fig. 5). The influence of the solvent polarity on the fluorescence was analysed (Fig. 6) and although no direct correlation could be observed, few interesting observations were recorded. A significant loss in the fluorescence intensity and shift in the emission maximum (Em) was evidenced when water (polarity index, PI=10.1, Em = 545nm) and Methanol (PI=5.1, Em = 538nm) were used as the solvent. Although the fluorescent intensity dropped when Hexane (PI=0.1, Em = 509nm) was used as the solvent, Em was observed to be well within the range of 500-520nm, exhibited by other solvents used in this study.
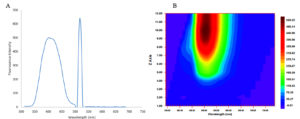
Fig. 5. (A) Shows the fluorescence spectral features of the partially purified fluorescent compound from Penicillium sp. MIP-3 (B) Contour graph showing the emission maximum.
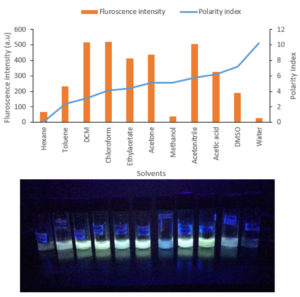
Fig. 6. (A) Graph summarises the solvatochromism analysis exhibited in different solvents. (B) The fluorescence exhibited in solvents was shown.
GC-MS analysis of the DCM extract of MIP-3 revealed the presence of 1-Buten-1-ol, 2-methyl-4-(2,6,6-trimethyl-1-cyclohexen-1-yl)-, formate, (1E)-, also known as Latia luciferin. In addition, steroids and steroid derivatives constituted to the majority of the compounds identified in the study (Table 1). β-Sitosterol accounted for (44.79%), Aspidinol (21.8%), Stigmasterol (9.33%), Campesterol (5.4%), p-Butyrophenetidide (3.05%), Benzo[h]quinoline, 2,4-dimethyl- (1.16%), Dodecanoic acid, 1,1′-biphenyl-4-yl carbonyl methyl ester (1.93%), 9H-Xanthene-9-carboxylic acid phenethyl amide (1.23%), l-Proline, and N-allyloxycarbonyl-, octyl ester (1.05%) were the major compounds identified from the mass spectrum (Fig. 7).
Table (1):
lists the major compounds observed in the GC-MS spectrum of Penicillium sp. MIP-3 extract.
RT |
Area% |
Library/ID |
CAS# |
|---|---|---|---|
15.65 |
1.05 |
l-Proline, N-allyloxycarbonyl-, octyl ester |
1000313-66-4 |
16.14 |
1.23 |
9H-Xanthene-9-carboxylic acid phenethyl amide |
349401-32-7 |
16.45 |
21.8 |
Aspidinol |
000519-40-4 |
16.59 |
1.93 |
Dodecanoic acid, 1,1′-biphenyl-4-yl carbonyl methyl ester |
004376-38-9 |
16.91 |
1.79 |
1-Buten-1-ol, 2-methyl-4-(2,6,6-trimethyl-1-cyclohexen-1-yl)-, formate, (1E)- |
021730-91-6 |
19.81 |
5.4 |
Campesterol |
000474-62-4 |
20.36 |
9.33 |
Stigmasterol |
000083-48-7 |
20.91 |
1.16 |
Benzo[h]quinoline, 2,4-dimethyl- |
000605-67-4 |
21.38 |
44.79 |
β-Sitosterol |
000083-46-5 |
22.16 |
3.05 |
p-Butyrophenetidide |
021218-92-8 |
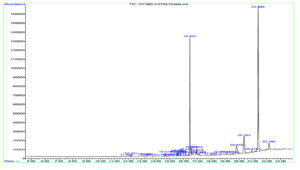
Fig. 7. Shows the GC-MS spectrum of Penicillium sp. MIP-3 extract.

Fig. 8. Shows the culture and spore (50x magnification) morphology of the fungal isolates on PDA. (A, B)Colletotrichum sp MIP-5 (C, D) Penicillium sp. MIP-3.
Identification of fungal isolates
The culture morphology and spore characteristics of the two fungal isolates were shown in Fig. 8. The MIP-5 isolate had a white cottony mycelium and produced rod shaped conidia with blunt ends, characteristic of Colletotrichum spp. Similarly, MIP-3 had a pale green mycelium with powdery mass of conidiospores borne on numerous erect sporangiophores, characteristic of Penicillium spp. The ITS sequences were amplified, sequenced and used to identify the fungal isolates. The blast search for the ITS sequence of MIP-3 (575bp) rendered Penicillium citrinum as the closest match with >98% sequence identity and 98% query coverage. The isolate MIP-3 resolved along with Penicillium citrinum in the maximum likelihood phylogenetic tree with 73% bootstrap support (Fig. 9). Similarly, the blast search for the ITS sequence of MIP-5 (548bp) rendered several Colletotrichum spp. as the closest match with 99.45% sequence identity and 100% query coverage. The isolate MIP-5 resolved along with C. queenslandicum, C. ti, C. arecicola, and C. tropicale and clade separation between them was not well supported (Fig. 9). Hence, it was identified only at the genus level to avoid erroneous naming. ITS sequences of the two isolates generated in the study was deposited in Genbank for global access under MZ955452 (Penicillium sp. MIP-3) and MZ959110 (Colletotrichum sp. MIP-5).
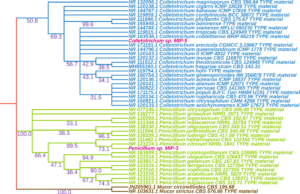
Fig. 9. Shows a maximum likelihood tree with bootstrap support values reflected on the nodes.
Endophytic fungi are widely recognized as source of novel bioactive secondary metabolites of significant biotechnology and therapeutic values.8,14 Intricate invasion and defence strategies of these cryptic organisms render them greater arsenal of metabolites to evade the plant defence, colonize and subsequently confer plant protection against pathogen and pest, in some cases confer physiological benefits like adaptation to heat, drought and salinity stresses.2
Taxol, claimed to be the first billion-dollar anticancer drug revolutionized the endophytic fungal research and exploration efforts grew exponentially.50 Taxol producers’ list spans across unrelated taxa and more interestingly were reported from diverse hosts that included both gymnosperms51-54 and angiosperms55,56 plants of varied ecosystems. Also, several intermediatory compounds and taxol derivatives collectively termed as Taxanes were reported from endophytic fungi with potential therapeutic values.57,58 Taxanes are used in palliative treatment of cancer, because of their anticancer activity and unique mechanism of action by stabilizing the microtubules.59 In the present study, presence of taxane in the fungal extracts were screened based on TLC plate by co-migrating them along with paclitaxel (Sigma-Aldrich). Among 31 endophytic fungal isolates, only five isolates had bands with similar Rf value of the paclitaxel and were considered as potential taxane producers. The spectral (UV-Visible and FTIR) and chromatographic (HPLC) data, suggested Colletotrichum sp. MIP-5 as a putative taxol producer. Despite the development of genetic screens for taxol, analytical methods were needed to ascertain the taxol production in real time. The primary reason being the temporary inactivation of the gene cassette due to the loss of environmental cues or permanent loss due to gene mutations or disruptions. Furthermore, the complexity of the biosynthetic pathway and lack of complete knowledge about the same impedes the genetic screening methods and analytical methods continue to garner wide support.60 Hence in this study, analytical methods were employed for ascertaining the putative taxol producers. Erstwhile, Colletotrichum genera has been reported to produce taxol55 and also a rich repertoire of other bioactive metabolites like 6-isoprenylindole-3-carboxylic acid, Colletotric acid, Colutellin A, Piperine, Asiaticoside, Quinine, Colletotrichine, Colletopeptides, Mellein, and Brevianamide F.61-68 The species boundaries in this cosmopolitan genera have been obscure and although considerable clarification have been reported through multilocus phylogeny,69 re-evaluation of the Colletotrichum taxonomy would be inevitable to ascertain the species identities.
Several fluorescent and bio-luminance molecules, primarily from mushrooms have been reported. A fluorescent molecule, ergosta-4,6,8 (14),22-tetraen-3-one, steroid derivative, was reported from Alternaria alternate.30,72 Territrems were lactone derivatives reported from Aspergillus terreus, exhibiting blue fluorescence (Em = 420nm).73 The mycotoxins from Penicillium janthinellum, Janthitrems belong to the Indole di-terpenoids class and have been reported to exhibit blue fluorescence (Em = 385nm).31,71 The fluorescence spectral feature (Em = 500-520nm) of the compound from our isolate Penicillium sp. MIP-3 overlapped with Flavin derivatives. A significant drop in the fluorescence intensity and shift in the Em was recorded when the compound from Penicillium sp. MIP-3 was dissolved in water (Em = 545nm) and methanol (Em = 538nm). Riboflavin (water soluble vitamin B2), a flavin derivative exhibiting greenish yellow fluorescence (emission range 500-600nm) was reported from several fungi and Ashbya gossypii was employed for its industrial production.74
GC-MS based metabolite profiling pointed out to the presence of Latia luciferin (1-Buten-1-ol, 2-methyl-4-(2,6,6-trimethyl-1-cyclohexen-1-yl)-, formate, (1E)-) in the extract of Penicillium sp. MIP-3. Latia luciferin had been implicated in the bio-luminescence (Em = 536nm) of a fresh water limpet Latia neritoides.75 Luciferins and their associated enzyme systems had been implicated in the bio-luminescence of fungi and other luminescent organisms.28,76 Recently, 3hydroxyhispidin, a fungal luciferin has been reported to be involved in the bio-luminescence of fungi.33 Harvey77 had reported the luminescence phenomenon accompanying the oxidation of luciferin without luciferase under oxidizing conditions or with gentle heating in alcohol. Recently, Yu et al.78 reported that the Latia luciferin was lacking fluorescence potential based on the theoretical studies of the spectral features, geometrical and electronic structures. Purification and further characterization of the fluorescent molecule from Penicillium sp. MIP-3 can shed more information. Nevertheless, this has been the first report of Latia luciferin from fungal source. In addition, DCM extract of Penicillium sp. MIP-3 had several biologically active steroid derivatives. β-Sitosterol, a prominent antioxidant, and anticholesteremic drug79 was the prominently observed metabolites. Recently, β-Sitosterol had been reported for its immunomodulatory effect against SARS-Cov2.80 The observance of sterol and fatty acid derivatives as the major compounds in GC-MS spectrum could be attributed to the mid-polar DCM extraction procedure employed. Aspidinol, a phloroglucinol derivative with antibacterial activity81 was also observed. Bioprospecting these organisms hold greater promise due to their ability to produce wide variety of novel and potential compounds.
In the present study, about 31 fungal isolates were recovered from the Mangifera indica leaves. Among five putative taxol producers identified based on TLC, Colletotrichum sp. MIP-5 was reported to be a potential producer of taxanes. Mass production and purification of the taxanes could help in elucidating the structural identity of the taxanes and subsequently could serve as a potential source of anticancer metabolites. Also, Penicillium sp. MIP-3 was observed to produce fluorescent molecule with Flavin like fluorescence spectrum. Based on GC-MS analysis, Latia luciferin was reported for the first time from this fungal source. In addition, therapeutically active steroid, quinones and phloroglucinol derivatives were also reported from Penicillium sp. MIP-3. These molecules could potentially be used in biological applications and needs further studies.
Additional file: Additional Table S1.
ACKNOWLEDGMENTS
The authors are thankful to the DST-PURSE and School of Chemistry, MKU for the instrumentation facilities. Authors profusely thank the anonymous reviewers for reviewing our manuscript.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
MP designed and guided the study. EGJ, PS conducted the experiments, analyzed the data and drafted the manuscript. SSK helped with phylogenetic analysis and manuscript preparation. MP reviewed the manuscript.
FUNDING
This study was funded by Science and Engineering Research Board (SERB), Goverment of India (SR/FT/LS-50/2011).
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
The sequences generated in the study were submitted in Genbank (MZ955452 and MZ959110, awaiting release) and were being processed for global release. All reference sequences employed in the study were retrieved from Genbank and their accession numbers were reflected in the phylogenetic tree and presented in supplementary file, S1.
- Arnold AE, Lutzoni F. Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology. 2007;88(3):541-549.
Crossref - Rodriguez RJ, White JF Jr, Arnold AE, Redman RS. Fungal endophytes: diversity and functional roles. New Phytol. 2009;182(2):314-330.
Crossref - Spiteller P. Chemical ecology of fungi. Nat Prod Rep. 2015;32(7):971-993.
Crossref - Scherlach K, Hertweck C. Mediators of mutualistic microbe-microbe interactions. Nat Prod Rep. 2018;35(4):303-308.
Crossref - Wongsuk T, Pumeesat P, Luplertlop N. Fungal quorum sensing molecules: Role in fungal morphogenesis and pathogenicity. J Basic Microbiol. 2016;56(5):440-447.
Crossref - Padder SA, Prasad R, Shah AH. Quorum sensing: A less known mode of communication among fungi. Microbiol Res. 2018;210:51-58.
Crossref - Strobel GA, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol Biol Rev. 2003;67(4):491-502.
Crossref - Suryanarayanan TS, Thirunavukkarasu N, Govindarajulu M, Sasse F, Jansen R, Murali TS. Fungal endophytes and bioprospecting: An appeal for a concerted effort. Fungal Biol Rev. 2009;23(1-2):9-19.
- Nisa H, Kamili AN, Nawchoo IA, Shafi S, Shameem N, Bandh SA. Fungal endophytes as prolific source of phytochemicals and other bioactive natural products: A review. Microb Pathog. 2015;82:50-59.
Crossref - Gouda S, Das G, Sen SK, Shin HS, Patra JK. Endophytes: A Treasure House of Bioactive Compounds of Medicinal Importance. Front Microbiol. 2016;7:1538.
Crossref - Gupta M, Shukla KK. Endophytic Fungi: A Treasure Trove of Novel Bioactive Compounds. In: Bioactive Natural products in Drug Discovery, eds. Singh J, Meshram V, Gupta M. Springer, Singapore. 2020:427-449.
Crossref - Ibrar M, Ullah MW, Manan S, Farooq U, Rafiq M, Hasan F. Fungi from the extremes of life: an untapped treasure for bioactive compounds. Appl Microbiol Biotechnol. 2020;104(5):2777-2801.
Crossref - Priti V, Ramesh BT, Singh S, et al. How promising are endophytic fungi as alternative sources of plant secondary metabolites? Curr Sci. 2009;97(4):477-478.
- Kusari S, Spiteller M. Are we ready for industrial production of bioactive plant secondary metabolites utilizing endophytes? Nat Prod Rep. 2011;28(7):1203-1207.
Crossref - Stierle A, Strobel G, Stierle D. Taxol and Taxane production by Taxomyces andreanae, an endophytic fungus of Pacific Yew. Science. 1993;260(5105):214-216.
Crossref - Staniek A, Woerdenbag HJ, Kayser O. Screening the endophytic flora of Wollemia nobilis for alternative paclitaxel sources. J Plant Interact. 2010;5(3):189-195.
Crossref - Kumaran RS, Muthumary J, Hur BK. Isolation and identification of an anticancer drug, taxol from Phyllosticta tabernaemontanae, a leaf spot fungus of an angiosperm, Wrightia tinctoria. J Microbiol. 2009;47(1):40-49.
Crossref - Chakravarthi BVSK, Das P, Surendranath K, Karande AA, Jayabaskaran C. Production of paclitaxel by Fusarium solani isolated from Taxus celebica. J Biosci. 2008;33(2):259-267.
Crossref - Yanyan Z, Ting H, Qianliang M, Lingshang W, Khalid R, Luping Q. Alkaloids Produced by Endophytic Fungi: A Review. Natural Product Communication. 2012;7(7):963-968.
Crossref - de Souza JJ, Vieira IJ, Rodrigues-Filho E, Braz-Filho R. Terpenoids from endophytic fungi. Molecules. 2011;16(12):10604-10618.
Crossref - Rabia T, Aqeel J, Yasir R. Fatty acids and their amide derivatives from endophytes: new therapeutic possibilities from a hidden source. FEMS Microbiol Lett. 2018;365(12):1-7.
Crossref - Manganyi MC, Ateba CN. Untapped Potentials of Endophytic Fungi: A Review of Novel Bioactive Compounds with Biological Applications. Mircroorganisms. 2020;8(12):1934.
Crossref - Deshmukh SK, Gupta MK, Prakash V, Saxena S. Endophytic Fungi: A Source of Potential Antifungal Compounds. J Fungi (Basel). 2018;4(3):77.
Crossref - Rahul CM, Mayurika G, Colin JB, Deshmukh SK. Endophytic Fungi – An Untapped Source of Potential Antioxidants. Current Bioactive Compounds. 2020;16(7):944-964.
Crossref - Deshmukh SK, Verekar SA, Bhave SV. Endophytic fungi: a reservoir of antibacterials. Front Microbiol. 2015;5:715.
Crossref - Uzma F, Mohan CD, Hashem A, et al. Endophytic Fungi – Alternative Sources of Cytotoxic Compounds: A Review. Front Pharmacol. 2018;9:309.
Crossref - Newman DJ, Cragg GM. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J Nat Prod. 2020;83(3):770-803.
Crossref - Purtov KV, Petushkov VN, Baranov MS, et al. The chemical basis of fungal bioluminescence. Angew Chem Int Ed Engl. 2015;54(28):8124-8128.
Crossref - Teranishi K. Identification of possible light emitters in the gills of a bioluminescent fungus Mycena chlorophos. Luminescence. 2016;31(7):1407-1413.
Crossref - Seitz LM, Paukstelis JV. Metabolites of Alternaria alternata: ergosterol and ergosta-4,6,8(14),22-tetraen-3-one. J Agric Food Chem. 1977;25(4):838-841.
Crossref - Gallagher RT, Latch GC, Keogh RG. The janthitrems: fluorescent tremorgenic toxins produced by Penicillium janthinellum isolates from ryegrass pastures. Appl Environ Microbiol. 1980;39(1):272-273.
Crossref - Abbas CA, Sibirny AA. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol Mol Biol Rev. 2011;75(2): 321-360.
Crossref - Kotlobay AA, Sarkisyan K, Mokrushina Y, et al. Genetically encodable bioluminescent system from fungi. Proc Natl Acad Sci USA. 2018;115(50):12728-12732.
Crossref - Mitiouchkina T, Mishin AS, Somermeyer LG, et al. Plants with self-sustained luminescence. BioRxiv. 2019.
- Khakhar A, Starker CG, Chamness JC, et al. Building customizable auto-luminescent luciferase-based reporters in plants. eLife. 2020;9:e52786.
Crossref - Taylor JW, Goker M, Pitt JI. Choosing one name for pleomorphic fungi: The example of Aspergillus versus Eurotium, Neosartorya and Emericella. Taxon. 2016;65(3):593-601.
Crossref - Slepecky RA, Starmer WT. Phenotypic Plasticity in Fungi: A Review with Observations on Aureobasidium pullulans. Mycologia. 2009;101(6):823-832.
Crossref - Seifert KA. Progress towards DNA barcoding of fungi. Mol Ecol Resour. 2009;9(Supl. S1):83-89.
Crossref - Schoch CL, Seifert KA, Huhndorf S, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A. 2012;109(16):6241-6246.
Crossref - Suryanarayanan TS, Kumaresan V, Johnson JA. Foliar fungal endophytes from two species of the mangrove Rhizophora. Can J Microbiol. 1998;44(10):1003-1006.
Crossref - Li J, Strobel G, Sidhu R, Hess WM, Ford EJ. Endophytic taxol-producing fungi from bald cypress. Microbiology. 1996;142(8):2223-2226.
Crossref - Gokul Raj K, Manikandan R, Arulvasu C, Pandi M. Anti-proliferative effect of fungal taxol extracted from Cladosporium oxysporum against human pathogenic bacteria and human colon cancer cell line HCT 15. Spectrochim Acta A Mol Biomol Spectrosc. 2015;138:667-674.
Crossref - Soloman RDJ, Santhi VS. Purification of bioactive natural product against human microbial pathogens from marine sea weed Dictyota acutiloba J. Ag. World J Microbiol Biotechnol. 2008;24(9):1747-1752.
Crossref - Mydeen SS, Kumar RR, Kottaisamy M, Vasantha VS. Biosynthesis of ZnO nanoparticles through extract from Prosopis juliflora plant leaf: Antibacterial activities and a new approach by rust-induced photocatalysis. Journal of Saudi Chemical Society. 2020;24(5):393-406.
Crossref - Popczyk A, Cheret Y, El-Ghayoury A, Sahraoui B, Mysliwiec J. Solvatochromic fluorophores based on thiophene derivatives for highly-precise water, alcohols and dangerous ions detection. Dyes and Pigments. 2020;177:108300.
Crossref - Konappa N, Udayashankar AC, Krishnamurthy S, Pradeep CK, Chowdappa S, Jogaiah S. GC-MS analysis of phytoconstituents from Amomum nilgiricum and molecular docking interactions of bioactive serverogenin acetate with target proteins. Sci Rep. 2020;10(1):16438.
Crossref - Cenis JL. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992;20(9):2380.
Crossref - White TJ, Bruns T, Lee S, Taylor JL. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: A Guide to methods and applications, eds. Innis MA, Gelfand DH, Sninsky JJ, White TJ. Academic Press, Inc., New York. 1990;315-322.
Crossref - Sundaresan N, Jagan EG, Kathamuthu G, Pandi M. Internal transcribed spacer 2 (ITS2) molecular morphometric analysis based species delimitation of foliar endophytic fungi from Aglaia elaeagnoidea, Flacourtia inermis and Premna serratifolia. PLoS ONE. 2019;14(4):e0215024.
Crossref - Flores-Bustamante ZR, Rivera-Orduna FN, Martinez-Cardenas A, Flores-Cotera LB. Microbial paclitaxel: advances and perspectives. J Antibiot. 2010;63(8):460-467.
Crossref - Garyali S, Kumar A, Reddy MS. Diversity and antimitotic activity of taxol-producing endophytic fungi isolated from Himalayan yew. Ann Microbiol. 2013;64(3):1413-1422.
Crossref - Strobel G, Yang X, Sears J, Kramer R, Sidhu RS, Hess WM. Taxol from Pestalotiopsis microspora, an endophytic fungus of Taxus wallachiana. Microbiol. 1996;142(Pt 2):435-440.
Crossref - Xiong Z, Yang Y, Zhao N, Wang Y. Diversity of endophytic fungi and screening of fungal paclitaxel producer from Anglojap yew, Taxus x media. BMC Microbiol. 2013;13:71-81.
Crossref - Zhou X, Wang Z, Jiang K, et al. Screening of taxol-producing endophytic fungi from Taxus chinensis var. mairei. Appl Biochem Microbiol. 2007;43(4):439-443.
Crossref - Gangadevi V, Muthumary J. Isolation of Colletotrichum gloeosporioides, a novel endophytic taxol-producing fungus from the leaves of a medicinal plant, Justicia gendarussa. Mycol Balcanica. 2008;4:1-4
- Yang Y, Zhao H, Barrero RA, et al. Genome sequencing and analysis of the paclitaxel-producing endophytic fungus Penicillium aurantiogriseum NRRL 62431. BMC Genomics. 2014;15:69.
Crossref - Parc G, Canaguier A, Landre P, Hocquemiller R, Chriqui D, Meyer M. Production of taxoids with biological activity by plants and callus culture from selected Taxus genotypes. Phytochemistry. 2002;59(7):725-730.
Crossref - Ottaggio L, Bestoso F, Armirotti A, et al. Taxanes from shells and leaves of Corylus avellana. J Nat Prod. 2008;71(1):58-60.
Crossref - Fauzee NJ. Taxanes: promising anti-cancer drugs. Asian Pac J Cancer Prev. 2011;12(4):837-851. PMID: 21790213
- Kusari S, Singh S, Jayabaskaran C. Rethinking production of Taxol® (paclitaxel) using endophyte biotechnology. Trends Biotechnol. 2014;32(6):304-311.
Crossref - Arivudainambi USE, Anand TD, Shanmugaiah V, Karunakaran C, Rajendran A. Novel bioactive metabolites producing endophytic fungus Colletotrichum gloeosporioides against multidrug-resistant Staphylococcus aureus. FEMS Immunol Med Microbiol. 2011;61(3):340-345.
Crossref - Chithra S, Jasim B, Sachidanandan P, Jyothis M, Radhakrishnan EK. Piperine production by endophytic fungus Colletotrichum gloeosporioides isolated from Piper nigrum. Phytomedicine. 2014;21(4):534-540.
Crossref - Ren, Y, Strobel GA, Graff JC, et al. Colutellin A, an immunosuppressive peptide from Colletotrichum dematium. Microbiol. 2008;154(7):1973-1979.
Crossref - Yang ZD, Li ZJ, Zhao JW, Sun JH, Yang LJ, Shu ZM. Secondary metabolites and PI3K inhibitory activity of Colletotrichum gloeosporioides, a fungal endophyte of Uncaria rhynchophylla. Curr Microbiol. 2019;76(7):904-908.
Crossref - Zou WX, Meng JC, Lu H, et al. Metabolites of Colletotrichum gloeosporioides, an Endophytic Fungus in Artemisia mongolica. J Nat Prod. 2000;63:1529-1530.
Crossref - Wei B, Yang ZD, Chen XW, et al. Colletotrilactam A-D, novel lactams from Colletotrichum gloeosporioides GT-7, a fungal endophyte of Uncaria rhynchophylla. Fitoterapia. 2016;113:158-163.
Crossref - Lu H, Zou WX, Meng JC, Hu J, Tan RX. New bioactive metabolites produced by Colletotrichum sp., an endophytic fungus in Artemisia annua. Plant Science. 2000;151(1):67-73.
Crossref - Andre A, Wojtowicz N, Toure K, Stien D, Eparvier V. New acorane sesquiterpenes isolated from the endophytic fungus Colletotrichum gloeosporioides SNB-GSS07. Tetrahedron Letters. 2017;58(13):1269-1272.
Crossref - Bhunjun CS, Phukhamsakda C, Jayawardena RS, Jeewon R, Promputtha I, Hyde KD. Investigating species boundaries in Colletotrichum. Fungal Diversity. 2021;107:107-127.
Crossref - Sabry S, Ghanem K, Ghozlan H. Riboflavin production by Aspergillus terreus from beet-molasses. Microbiologia. 1993;9:118-124.
- Babu JV, Popay AJ, Miles CO, Wilkins AL, di Menna ME, Finch SC. Identification and Structure Elucidation of Janthitrems A and D from Penicillium janthinellum and Determination of the Tremorgenic and Anti-Insect Activity of Janthitrems A and B. J Agric Food Chem. 2018;66(50):13116-13125.
Crossref - Quang DN, Bach DD. Ergosta-4,6,8(14),22-tetraen-3-one from Vietnamese Xylaria sp. possessing inhibitory activity of nitric oxide production. Nat Prod Res. 2008;22(10):901-906.
Crossref - Ling KH, Yang CK, Peng FT. Territrems, tremorgenic mycotoxins of Aspergillus terreus. Appl Environ Microbiol. 1979;37(3):355-357.
Crossref - Aguiar TQ, Silva R, Domingues L. Ashbya gossypii beyond industrial riboflavin production: a historical perspective and emerging biotechnological applications. Biotechnol Adv. 2015;33(8):1774-1786.
Crossref - Shimomura O, Johnson FH. Purification and properties of the luciferase and of a protein cofactor in the bioluminescence system of Latia neritoides. Biochemistry. 1968;7(7):2574-2580.
Crossref - Wang MY, Liu YJ. Chemistry in Fungal Bioluminescence: A Theoretical Study from Luciferin to Light Emission. J Org Chem. 2021;86(2):1874-1881.
Crossref - Harvey EN. Studies on the oxidation of luciferin without luciferase and the mechanism of bioluminescence. Journal of Biological Chemistry. 1928;78:369-375.
Crossref - Yu M, Ohmiya Y, Naumov P, Liu YJ. Theoretical Insight into the Emission Properties of the Luciferin and Oxyluciferin of Latia. Photochem Photobiol. 2018;94(3):540-544.
Crossref - Rudkowska I, AbuMweis SS, Nicolle C, Jones PJ. Cholesterol-lowering efficacy of plant sterols in low-fat yogurt consumed as a snack or with a meal. J Am Coll Nutr. 2008;27(5): 588-595.
Crossref - Khan SL, Siddiqui FA. Beta-Sitosterol: As Immunostimulant, Antioxidant and Inhibitor of SARS-CoV-2 Spike Glycoprotein. Arch Pharmacol Ther. 2020;2(1):12-16.
Crossref - Hua X, Yang Q, Zhang W, et al. Antibacterial Activity and Mechanism of Action of Aspidinol Against Multi-Drug-Resistant Methicillin-Resistant Staphylococcus aureus. Front Pharmacol. 2018;9:619.
Crossref - Okwu DE, Ezenagu V. Evaluation of the phytochemical composition of Mango (Mangifera indica Linn) stem, bark and leaves. Int J Chem Sci. 2008;6(2):705-716
- Wu JB, Zhan RL, Liu F, Li GP, Zhao YL, Chang JM. Isolation, identification and diversity analysis of endophytic fungi in Mangifera indica. Journal of Fruit Science. 2013;30(6):1030-1039
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.