ISSN: 0973-7510
E-ISSN: 2581-690X
Sponge is an invertebrate animal in the Porifera phylum, which potentially produces bioactive compounds that are known as potential agents in producing antimicrobial compounds. This research aims to determine the antimicrobial activity of bacteria isolates obtained from the samples of the sponge of Kodingareng Keke Island-Makassar Indonesia. Samples were isolated by the pour plate method using NA (Nutrient Agar) media and incubated for 1 x 24 hours. A total of 8 bacteria were isolated with code KDR-02-01, KDR-02-02, KDR-02-03, KDR-02-04, KDR-02-05, KDR-02-06, KDR-02-07, KDR-02-08, KDR-02-09 and KDR-02-10. Each isolate of bacteria is purified by using quadrant streaks method until it is obtained by a pure isolate bacteria followed by an antagonistic test. From the antagonist test, bacteria KDR-02-07 continue to be fermented because they have the best inhibition compared to other isolates capable of inhibiting E. coli, S. aureus and C. albicans. Active isolate fermented for 8 days to produce secondary metabolite. After fermentation of 8 days the fermentation medium was separate by filtration then supernatant was extraction using ethyl acetate (1:1) and biomass macerated using methanol. Ethyl acetate extracts and methanol extracts that have been obtained are continued on antimicrobial activity test with diffusion method in order to microbial test microbes E. coli, S. aureus, B. subtilis, C. albicans and A. niger. The results of the study showed that ethyl acetate extract in concentration of 10% can inhibit E. coli and B. subtilis, in concentration of 5% inhibit B. subtilis and in concentrations of 1.25% inhibit A. niger, whereas in methanol extracts not acquired antimicrobial activity. Based on the results of a microscopic test it was obtained that the bacteria were isolated is Gram-positive and basil bacteria.
Antimicrobial, Sponge, bacteria, Gram positive
Indonesia has a very large marine biodiversity, including microbes, plants and animals. Indonesian Territory conditions in the form of archipelago, maritime and a supportive tropical climate, making Indonesia rich in biodiversity.1 Several studies have shown that marine life has enormous potential in producing bioactive compounds that can be used as medicinal raw materials. One of the marine biota that is known to produce these active compounds is sponges2,3 and among the many marine biota, sponges occupy the top place as a source of active substances.4
Sponges include invertebrate animals in the phylum porifera, which has the potential to produce bioactive compound.5 This is evident from more than 6000 bioactive substances that have been isolated from marine biota during the last three decades, and 40% of them were isolated from sponges.6,7
It is suspected that the diversity and presence of high bioactive compounds in sponges is related to the association of bacteria that are abundant in marine sponges The number of bacteria were found in sponges is around 40-60% of the total sponge biomass.8 Marine microorganisms are good candidates for new pharmaceuticals and bioactive natural products. There is accumulating evidence that demonstrates the involvement of symbiotic microorganism in the natural products originally attributed to the sponge host. Numerous natural products from marine invertebrates show striking structural similarities to metabolites of microbial origin, suggesting that microorganisms are the true source of these metabolites or are intricately involved in their biosynthesis.9,10
A total of 133 bacterial isolates were recovered from 55 sponges, a Demospongie class. Ten of isolates are produce active secondary metabolites as antibacterial agents against Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, Bacillus subtilis, Salmonella typhi and Escherichia coli.11 The Actinobacteria isolate was isolated from sponges producing secondary metabolites against S. aureus antibiotic resistant.12
The use of bacteria that live in symbiosis with sponges in the form of symbionts is better because they can be purified and cultured on a laboratory scale so that they do not need to be collected from nature and can be used as raw materials for producing antimicrobial compounds to fight infectious diseases.10 The aim of research are isolation of bacteria associated with sponges that have antibacterial activity.
Sampling technique
The sponges sample in this research are sponges from Kodingareng Keke island located in the Spermonde archipelago group, Makassar City, South Sulawesi Indonesia. The sponges were taken at shore water area at a depth 7-10 m below sea level by SCUBA Diving. The sponges was put into the coolbox and the brought to the laboratory
Isolation of bacteria from sponge samples
The sponge samples were washed with the sterile water then cut into small pieces approximately 1g of each sponge sample ground with a sterile mortar and pestle. Aliquots (0.1ml) were 10-fold diluted (10-3, 10-4 and 10-5) in sterile filtered seawater and 1 ml aliquots put into the petri dish and was added 15 ml NA medium in sea water then homogenized. Plates were incubated at 37°C for 2-3 days. Bacterial colonies were purified by transferring onto new plates for further use.
Antagonistic Assay
Pure culture of bacteria isolates were evaluation of antimicrobial activity by antagonistic assay using test microbes were taken from Laboratory of Microbiology Faculty of Pharmacy including Escherichia coli, Staphylococcus aureus, Bacillus subtilis, Candida albicans and Aspergillus niger. A small agar block of bacteria isolate were cutted using sterile stainless cylinder borer, then putted on the NA medium for bacteria and PDA medium for fungi that containing test microbes. The clear zone around the agar block after incubation periods were considered as active isolates.13
Fermentation and Extraction of antimicrobial substances
Isolates that were active in the antagonist test were made pre-culture (starter) in 500 mL Erlenmeyer flask containing 200 mL of NB (Nutrient Broth) as a fermentation medium. Fermentation was carried out at room temperature for 8 days by agitation condition with rotary at 150 rpm. After fermentation, biomass and supernatant was separated by filtration. Supernatant was extracted with ethyl acetate (1:1) using separating funnel, whereas biomass was extracted with methanol by maceration. The liquid extract was evaporated by rotary evaporation then the dried extract were determined antimicrobial activity.
Antimicrobial activity
Metahol and ethyl acetate extract were determined antimicrobial activity by agar diffusion method with serial concentration 1.25; 2.5; 5; 10%. A total 20µL of each extract concentration was dripped on paper disc. The disc paper were placed on the NA (Nutrient Agar) medium surface that had inoculated with test bacteria, and PDA (Potato Dextrose Agar) medium for fungi. Inhibitory zone were measured after incubation at 37°C for 24h for bacteria and 72h at room temperature for fungi.
Gram Staining
Observations were made using a microscope by placing on a purified object 1 glass culture which was then fixed. Then Gram was done by using violet crystal dye (gram A), iodine (gram B), acid alcohol (gram C), and safranin (gram D) and then allowed to stand for a few minutes. Rinse slowly then observe on a light microscope at 100x objective magnification.
Isolation of Symbiont Bacteria
The samples of sponges used in this study were ten sponges which were coded KDR-02-01, KDR-02-02, KDR-02-03, KDR-02-04, KDR-02-05, KDR-02-06, KDR. -02-07, KDR-02-08, KDR-02-09 and KDR-02-10 originating from the Kodingareng Keke Island Makassar Indonesia (Fig. 1).
The isolation of symbiont bacteria from sponges was carried out by pouring method using NA medium dissolved in sea water. From 10 sponge samples, 8 bacterial isolates were obtained from 8 sponges (Fig. 2).
Antagonist assay
Eight bacterial isolates were tested antagonistically using agar block method to screening their antimicrobial activity. The result revealed that isolate KDR-02-07 had better activity than the isolate KDR-02-04, KDR-02-01, KDR-02-05, KDR-02-6, KDR-02-08, KDR-02-09, KDR-02-03 to inhibit the E.coli, B.subtilis and C.albicans. All isolate were not inhibit A.niger (Table1).
Table (1):
Antagonistic test of symbiont sponge bacteria.
| Isolate Code | Microbes test | ||||
|---|---|---|---|---|---|
| E.coli | S.aureus | B.subtilis | C.albicans | A.niger | |
| KDR-02-04 | + | + | – | – | – |
| KDR-02-01 | – | – | – | – | – |
| KDR-02-05 | + | – | – | – | – |
| KDR-02-06 | ++ | – | + | – | – |
| KDR-02-07 | +++ | – | +++ | +++ | – |
| KDR-02-09 | ++ | – | – | + | – |
| KDR-02-03 | – | – | +++ | – | – |
| KDR-02-08 | + | – | + | – | – |
(-): no inhibition zone; (+) : diameter inhibition zone 1-5 mm;
(++): diameter inhibition zone 6-10 mm
(+++): diameter inhibition zone 11-15 mm
Fermentation of Active Isolate
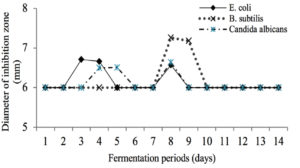
Active isolate KDR-02-07 from the antagonistic test then fermentation to produce the secondary metabolite. Fermentation was carried out using pure isolates which had been rejuvenated for 24h at 37°C. The fermentation process was carried out using Nutrient Broth (NB) medium. The determination of the fermentation periods was based on the results of the activity test of the fermentation liquid which was sampled for 14 days against the tested microorganisms. The results of the activity test showed that the 8th day fermentation showed the highest activity in inhibiting the test microbes so that the next fermentation process was carried out for 8 days.
After fermentation for 8 days, the supernatant and biomass were separated by filtration. The supernatant was extracted with ethyl acetate. Ethyl acetate is used because it is a semipolar solvent, does not mix with water and can extract more secondary metabolites which are antibacterial biomass was extracted with methanol by maceration.
Fig. 3. The Effect of fermentation time of KDR-02-07 isolates (days) on the diameter of inhibition zone (mm).
Antimicrobial Activity Test
The antimicrobial activity test was carried out by the agar diffusion method using a paper disk with a diameter of 6 mm. The test microbes used were Escherichia coli, Stapyhlococcus aureus, Bacillus subtilis, Candida albicans and Aspergillus niger tests. The diffusion method should be used because this method has the advantage of being simple and easy to interpret the results obtained.14 The results of the antimicrobial activity test of ethyl acetate and methanol extracts showed that the two extracts could not inhibit the growth of the tested fungi (C. albicans and A. niger). Ethyl acetate extract could only inhibit the growth of E.coli and B.subtilis bacteria, while methanol extract could not inhibit the test microbes. The ethyl acetate extract could not inhibit S. aureus bacteria, this was due to the morphological differences between the two Gram positive bacteria (S. aureus and B. subtilis). S. aureus has a cocci morphology while B. subtilis has a bacil morphology, so the surface area of the bacteria is different. According to Hatsock (2003) the size of bacterial cells affects the ability of cells to absorb compounds.15
Identification of Isolate KDR02-07
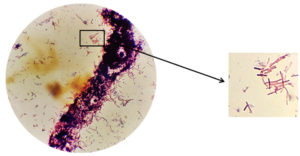
The identification of symbiont bacteria was carried out using a Gram staining procedure. The Gram staining procedure was chosen because two kinds of observation can be carried out in one time, namely observing the morphological shape of the bacteria and observing the type of bacteria tested, namely Gram-negative or Gram-positive. The gram staining procedure is carried out using Gram A (Crystal violet) dye which is purple which functions to give color to the target microorganism, Gram B (iodine) which functions to fix the primary paint which is absorbed by the target microorganism, gram C (ethanol) which functions to dissolve the previous stain and Gram D (safranin) which is red which is a secondary stain. Before stain, the symbiont bacteria isolate, which will be morphologically identified, must first be made preparations and then dripped with stain.
Based on the results of microscopic observations using the gram stain procedure revealed that the bacteria KDR-02-07 identified as Gram-positive bacteria with a bacil form. This can be seen from the observation under a microscope that shows the bacteria are purple. Gram-positive bacteria have a thick cell wall, a layer of cell membrane, and do not have an outer membrane. Gram positive bacteria will absorb and retain the crystal violet color which will be strengthened by iodine and will not be washed away after being decolorized by alcohol, therefore the color of Gram positive bacteria observed is blue or purple under a microscope.16
Table (2):
Antimicrobial activity of ethyl acetate and methanol from symbiont bacteria KDR-02-07.
| Concentration of extract | Extract | Inhibition zone diameter (mm) | ||||
|---|---|---|---|---|---|---|
| E.coli | S.aureus | B.subtilis | C.albicans | A.niger | ||
| 1,25% | Ethyl acetate | – | – | – | – | – |
| methanol | – | – | – | – | – | |
| 2,5% | Ethyl acetate | – | – | – | – | – |
| methanol | – | – | – | – | – | |
| 5% | Ethyl acetate | – | – | 6,41 | – | – |
| methanol | – | – | – | – | – | |
| 10% | Ethyl acetate | 6,61 | – | 6,69 | – | – |
| methanol | – | – | – | – | – | |
| Positive control | Ampicillin | 15,29 | 10,54 | 15,48 | – | – |
| Nystatin | 15.55 | 12.23 | ||||
| Negative control | – | – | – | – | – | – |
Thin Layer Chromatography Profile
The TLC profile of ethyl acetate and methanol extract had different compound profiles, this indicated that the two extracts contain different compounds whereas ethyl acetate extract contain more compound than methanol extract. That way the methanol extract could not inhibit the microbe test.
Ten sponges sample obtained from Kodingareng Keke island, 8 bacteria were isolated and bacteria KDR02-07 were able to inhibit the growth of E.coli, B.subtilis. The strongest antibacterial activity was shown at a concentration of 5% and 10% of the ethyl acetate extract against B. subtilis, and a concentration of 10% against E. coli, while the methanol extract could not inhibit the test microbes used. Base on Gram staining revealed that bacteria KDR02-07 was Gram positive and bacil bacteria.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This research was supported by Basic Research Grants from Ministry of Research and Technology, Indonesia with Grant Number: 7/AMD/EI/KP.PTNBH/2020.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Friedrich AB, Fischer I, Proksch P, Hacker J, Hentschel U. Temporal variation of the microbial community associated with the mediterranean sponge Aplysina aerophoba. FEMS Microbiol Ecol. 2001;38(2-3):105-113.
Crossref - Muniarsih T, Satari RR. Isolation of Antimicribial bioactive substances from sponges From Pari Island, Seribun . Proceedings of Marine Biotechnology Seminar Indonesia Prosidings. Indonesian Institute of Sciences, Jakarta.1998:151-15
- Bharathiraja S, Rajasekaran R, Suriya J. Antibacterial activity of marine bacteria isolated from sponge Spirastrella inconstans. J Chem Pharm Res. 2014;6(11):395-398
- Libes S. An Introduction to Marine Biogeochemistry. John Wiley & Sons, New York. 1992
- Taylor MW, Radax R, Steger D, Wagner M. Sponge-Associated Microorganisms : Evolution, Ecology, and Biotechnological Potential. Microbiol Mol Biol Rev. 2007;71(2):295-347.
Crossref - Imhoff JF, Stohr R. Sponge associated bacteria: general overview and special aspect of the diversity of bacteria associated with Halichondria panicea. Mol Biotechnol. 2003;1:35-57.
Crossref - Engel S, Jensen PR, Fenical W. Chemical ecology of marine microbial defense. J Chem Ecol. 2002;28(10):1971-1985.
Crossref - Abubakar H., Wahyudi AT, abd Yuhana M. Skrining Bakteri yang Berasosiasi dengan Spons Jaspis sp . Sebagai Penghasil Senyawa Antimikroba. Indonesian Journal of Marine Sciences. 2011;16(1):35-40.
- Zheng L, Han X, Chen HM, Lin W, Yan XJ. Marine bacteria associated with marine macroorganisms: the potential antimicrobial resources. Ann Microbiol. 2005;55(2):119-124.
- Lee YK, Lee JH, Lee HK, Microbial Symbiosis in Marine Sponges. J Microbiol. 2001:254-264.
- Trianto A, Radjasa OK, Sabdono A, et al. Exploration Culturable Bacterial Symbionts of Sponges From Ternate Island, Indonesia. Biodiversitas. 2019;20(3):776 – 782.
Crossref - Rante H, Wahyono, Murti YB, Alam G. Purification and Characterization of Anti-Multidrug resistances bacteria from actinomycetes associated sponge. Indonesian Journal of Pharmacy. 2010;21(3):158-165
- Rante H, Yulianty R, Evary YM, Hardiana E. Isolation and Antibacterial Activity of Endophytic Fungi from Melochia umbellata (Houtt). J Pure Appl Microbiol. 2017;11(3):1313-1318.
Crossref - Balouiri M, Sadiki M, Ibnsouda SK. Methods for In Vitro Evaluating Antimicrobial Activity: A Review. Journal of Pharmaceutical Analysis. 2016;6(2);71-79.
Crossref - Hartsock A. Bacterial Cytoplasm & Cell Membrane : Structure and Component. 2003. https://study.com/academy/lesson/bacterial-cytoplasm-cell-membrane-structure-components.html
- Elliot T, Whorthington. Medical Microbiology and Infection. EGC, Jakarta, 2013
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.