ISSN: 0973-7510
E-ISSN: 2581-690X
The interesting application of bile salt hydrolase enzyme is reduction of cholesterol in serum and amelioration lipid profile. While uricase enzyme can be applied to convert insoluble uric acid to be soluble form and excrete from the body. Probiotics are living organisms with generally know that they can provide beneficial effects to their host. Several reports show that probiotic bacteria with bile salt hydrolase and uricase can improve hypercholesterolemia and hyperuricemia patient. The novel isolate of Lactobacillus from Pak Sian Dong in this study is identified as L. brevis SF121 and probably use as probiotic bacteria in the future. However, this isolate still need further experiments to investigate and improve properties of probiotics. Moreover, this finding suggests that Pak Sian Dong or fermented spider plant can be designated as a good source for probiotic screening and also defines as health-promoting diet.
Lactobacillus, bile salt hydrolase, uricase, spider plant
Lactobacillus is gram positive bacteria in order Lactobacillales and commonly are defined as lactic acid producing bacteria based on their major by-product. This bacteria normally can be found in the environment, dairy product, animal gastrointestinal tract and fermented food.
Several studies have been reported that fermented food can be appropriated as the sources of lactic acid bacteria and also probiotics1-10. Probiotic bacteria with good properties are usually isolated from fermented vegetable, such as kimchi11 and sauerkraut12,13.
Pak Sian Dong is a kind of fermented vegetable in Thailand, which made from spider plant. Spider plants are affiliated with Cleomaceae family and designated the scientific name as Cleome gynandra L. These plants usually are observed worldwide, including Thailand. Fermentation can degrade toxic substance, called hydrocyanic which is defined as a neurotoxin, in spider plant then this kind of food will be safe to consume14.
Probiotics are assigned as living organism such as bacteria, mold and yeast, which provide advantages to their living host. The first screening criteria for probiotic bacteria are colonizing properties, which are able to attach the surface of the host and can survive in a gastrointestinal tract environment. The other criteria are beneficial properties that promote the health status of the host15-17.
Deconjugation of bile acid is occurred by catalytic enzyme calls bile salt hydrolase. Various bacteria have ability to produce this enzyme, especially lactic acid bacteria and Enterobacteria. The interesting application for bile salt hydrolase is cholesterol lowering. There are many reports that used bile salt hydrolase producing bacteria for decreasing of cholesterol level in blood of animal models and human volunteers18-20.
Gout is a disease which links to serum uric acid. Resulting of solubility of uric acid in blood is low, high level of this substance can cause precipitation of uric acid crystal leading to inflammation of joints. Therapeutic of this disease is probably aiming at; first, reduction of uric acid production by using allopurinol drug for xanthine oxidase activity inhibition; second, using probenecid to inhibit tubular reabsorption of uric acid and; third, using pegloticase for acting as uricase enzyme21,22. Uricase or urate oxidase enzyme has ability to transform insoluble uric acid to allantoin which is soluble substance and can be excreted by the kidney. Therefore, catalytic for uric acid activity, uricase is interested to be used for hyperuricemia and gout. Variety genus of bacteria have been screened and isolated with uricase activity, such as Bacillus23, 24, Pseudomonas25 and Lactobacillus26.
The objective of this study was the determination of bile salt hydrolase activity, uricase activity and colonizing properties in Lactobacillus brevis which was isolated from Pak Sian Dong in Pathum Thani, Thailand.
Strain isolation and cultivation conditions
Lactobacillus brevis SF121 was isolated from Pak Sian Dong. Pak Sian Dong were purchased from local area in Pathum Thani province, a gram of each sample was mixed with 9 ml of NaCl, 0.9% (w/v) or normal saline solution (NSS). The mixture was cultured on MRS agar which contained CaCO3 0.5% (w/v) and the plates were incubated for 48 h at 37⁰C incubator. Clear zone from CaCO3 degradation was observed around the colony and the colony was picked to determine for lactic acid bacteria characteristics. The characteristics were included gram reaction with positive results and catalase test with negative results. Blood hemolytic zone of the isolate was also observed after 37°C incubated at least 72 h. All bacterial isolates were named which including L. brevis SF121 and stored at -80°C in glycerol until used.
Bile salt hydrolase activity and uricase activity
For determination of bile salt hydrolase activity of the isolates, MRS medium with 0.5% (w/v) of glycodeoxycholic acid (GCDA) (SigmaAldrich, USA) or taurodeoxycholic acid (TDCA) (Sigma-Aldrich, USA) were prepared as described by Yasiri et al.27. The isolates were streaked on GDCA- or TDCA-MRS agar plate and then placed at 37°C for 48 – 72 h. After incubation, colony with halozone was observed and recorded for ability to produce bile salt hydrolase.
To determine uricase activity, plate assay with modification was performed23, 25. The isolate was streaked on MRS agar containing 0.5% (w/v) uric acid (Sigma-Aldrich, USA) and clear zone were observed after incubation for 48 – 72 h at 37°C.
Strain identification
API50CHL (Biomerioux, France) was used to identify the strain. In addition, the 16s rRNA sequence analysis also was used to confirm species of the strain. The bacterial genomic DNA of the isolate was extracted with GF-1 bacterial DNA extraction kit (Vivantis, Malaysia) and amplification of 16s rRNA by PCR was performed. The forward universal primer F8 (5’-AGAGTTTGATCCTGGCTCAG-3’) and reverse universal primer R1492 (5’-GGTTACCTT GTTACGACTT-3’) were used for amplification. After PCR amplification and agarose gel electrophoresis, Novel Juice (GeneDireX, USA) was mixed with the product and then observed the result on 0.8% (w/v) agarose (Invitrogen, USA). The PCR products were sequenced and the sequencing results were compared with a database (NCBI).
Gastric fluid and bile salt tolerance
To determine the tolerance of gastric fluid and bile salt, the procedure with slightly modified from Mota et al. was followed28. Synthetic gastric fluid was prepared as follows; weighed NaCl for 2 g and pepsin for 3.2 g and then mixed in 1000 ml of water. Added concentrated hydrochloric acid to solution to make the pH 2.5 and finally the solution was sterilized by filtration with 0.22 µm pore size membrane. To determine gastric fluid tolerance, bacterial suspension was prepared by washing and adjusting the overnight culture of L. brevis SF121 with NSS to McFarland standard no. 0.5. A hundred microliter of the L. brevis SF121 cell suspension was applied to 900 µl of synthetic gastric fluid and incubated for 3 h at 37°C. NSS was used instead of synthetic gastric fluid as a control group. The bacterial survival was evaluated and calculated on MRS agar by drop plate technique.
For bile salt tolerance test, liquid MRS with 0.3% (w/v) of bile salt was prepared and inoculated with 1% inoculum of L. brevis SF121 overnight culture. The numbers of bacteria were determined with the plate count technique at 0 h and 24 h of incubation. The percentage of survival after 24 h in bile containing condition was evaluated to assess bile salt tolerance.
Hydrophobicity
Hydrophobicity at cell surface of L. brevis SF121 was determined as following. Briefly, the overnight culture of L. brevis SF121 was washed twice with NSS and centrifuged to harvest the cells. The pellet of bacteria was adjusted to O.D. 0.4 – 0.5 at 600 nm (A0) with NSS. The bacterial suspension from A0 was pipet to test tube for 1.2 ml and mixed with 0.2 ml of xylene. After mixing, O.D. 600 nm measurement of a lower aqueous phase was performed. Calculation of hydrophobicity percentage by using (1-A1/A0) x 100 formula28.
Antibiotic susceptibility
In this study, the MIC of antibiotics, including, ampicillin, chloramphenicol, ciprofloxacin, erythromycin, nalidixic acid, penicillin, tetracycline and vancomycin were determined for antibiotic susceptibility following CLSI (Clinical and laboratory standards institute) guideline. In brief, contained antibiotic MRS broth was transferred to a microplate with a serial two-fold diluted from 512 µg/ml to 1 µg/ml. The overnight culture of L. brevis SF121 was diluted with NSS to prepare the suspension equal turbidity to 0.5 McFarland. The prepared L. brevis SF121 suspension was diluted hundred-fold with MRS and transferred to a filled microplate. The MIC was observed and interpreted after overnight incubation at 37°C.
Antimicrobial activity
The well diffusion assay was used to assess antimicrobial activity of L. brevis SF121. The twenty milliliters of Mueller-Hinton agar plates were prepared and swabbed with the indicator organisms. The indicator organisms in this study, including gram positive bacteria (Bacillus cereus, Staphylococcus aureus, Streptococcus pyogenes, Enterococcus faecium) gram negative bacteria (Escherichia coli, Pseudomonas aeruginosa, Salmonella enterica Typhimurium, Shigella sonnei, Vibrio cholerae) and yeast (Candida albicans). All organisms were cultivated in Mueller-Hinton broth at 37°C for overnight before spreading on agar plates. A well on swabbed plate that made from cork borer no.3 was filled with 100 µl of L. brevis SF121 overnight culture supernatant, which was prior adjusted the pH to 7.0 with 1 N NaOH and filtered by 0.22 µm pore size membrane for sterilization. The plates were measured for the inhibition zone after overnight incubation at 37°C.
Lactobacillus isolation and characterization
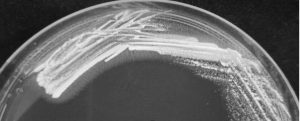
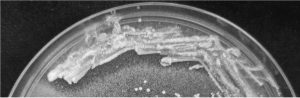
Gram positive rod shaped bacteria with catalase negative and no hemolytic activity was selected from MRS agar. Bile salt hydrolase activity was shown as precipitated halozone around the colony of L. brevis SF121 on GDCA-MRS agar plate (Fig. 1) but no activity observed on TDCA-MRS. The isolate also exhibited ability to convert insoluble uric acid in MRS agar plate to soluble form which showed as the clear zone around colony in the Fig. 2.
Fig. 1. The L. brevis SF121 on GDCA-MRS medium. The activity of bile salt hydrolase showed with halozone surrounding colonies.
Fig. 2. Uricase activity of L. brevis SF121 presented by clear zone on MRS medium containing uric acid.
Lactobacillus identification
The selected isolate showed 96.6% identity with Lactobacillus brevis by API 50 CHL. The identification was confirmed by 16s rRNA sequencing, which indicated coherent result with 99% identical to L. brevis in NCBI database. From isolation and identification, this isolate was designated as Lactobacillus brevis SF121.
Determination of probiotic properties
From general probiotic property characterization, gastric fluid tolerance of the isolate in this study was 67.07 % and bile salt tolerance was 89.62%. The L. brevis SF121 presented 21.44% hydrophobicity
For antibiotic susceptibility test, the isolate demonstrated sensitivity to only erythromycin and presented resistance to remaining antibiotics in this study (Table 1). Furthermore, all test organisms in the experiment, including gram positive bacteria, gram negative bacteria and yeast, could not be inhibited the growth by L. brevis SF121.
Table (1):
Antibiotic susceptibility profiles of L. brevis SF121 isolate.
| Antibiotics (µg/ml) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Ampi | Chlo | Cipr | Eryt | Nali | Peni | Tetr | Vanc | |
| L. brevis SF121 | 32 (Resist) |
8 (Resist) |
64 (Resist) |
0.5 (Sens) |
>256 (Resist) |
32 (Resist) |
16 (Resist) |
>256 (Resist) |
The antibiotics in this study consisted of ampicillin (Ampi), chloramphenicol (Chlo), ciprofloxacin (Cipr), erythromycin (Eryt), nalidixic acid (Nali), penicillin (Peni), tetracycline (Tetr) and vancomycin (Vanc).
From screening and isolation result, Lactobacillus species had been isolated from Pak Sian Dong. Previous study showed that the various genus of lactic acid bacteria can be found in Pak Sian Dong, including Lactobacillus sp. 29, thus the one report reveals potential of lactobacilli from this source have capability to use as probiotic 27. There was no hemolytic activity presented on blood agar plate which could be confirmed safety of the isolate for further used in animal and human17.
Determination of colonizing properties revealed that the isolate can resist simultaneous gastrointestinal environment. Both gastric fluid tolerance and bile salt tolerance are key important properties for survival of probiotic bacteria that appropriate for oral route use30. The properties confirm that this bacteria can survive and provide beneficial effects to the host at gastrointestinal tract region. For hydrophobicity, the isolate showed a little percentage, however the bacteria probably use other mechanisms to attach the epithelium cells such as aggregation property31.
The novel isolate has been identified as L. brevis, which commonly be found in milk, plants and fermented products. This bacteria can be applied to use in industrial and food technology and normally defined as GRAS (Generally Recognized As Safe)32. Some strain of L. brevis presented probiotic properties, e.g., antioxidant properties33, anti-caries activity34, immunomodulating activity35 and synergistic with prebiotic for antimicrobial activity36.
From the result of bile salt hydrolase, the isolate showed the activity only on MRS agar containing GDCA. This finding conformed to previous study that lactobacilli from Pak Sian Dong have an ability to hydrolyze GDCA but not TDCA27. According to bile salt hydrolase activity, this isolate can probably be alternatively used for cholesterol lowering application. Several reports published that lactobacilli, such as
L. plantarum37-39, L. gasseri40, L. johnsonii41,42 and L. brevis43, could be proved for bile salt hydrolase production. In 2013, L. plantarum with bile salt hydrolase was fed in high-cholesterol diet-induced rat could improve lipid profile in serum and liver of the rat44. Another report revealed that lactobacilli with strong expression of bile salt hydrolase have the capability to ameliorate hypercholesterolemia in mouse model20. In accordance with previous evidences, the novel strain, L. brevis SF121, might be able to reduce cholesterol by bile salt hydrolase activity.
Because of uricase enzyme is related to hyperuricemia therapy, this isolate with uricase activity is possibly appropriated to apply in this therapeutic approach. In human, uricase cannot be produced because of the loss of this enzymatic activity by genetic mutation during Miocene epoch45. Therefore, the compensation of this enzyme with intestinal microbes might be an important point. The normal route of administration of uricase (Pegloticase) is intravenous but there is a report that oral uricase administration in animal showed elimination of uric acid46,47. Moreover, alteration of Lactobacillus genus show relation with hyperuricemia in animal model48 so the adjustment of lactobacilli population probably a helpful for treatment.
L. brevis SF121, which isolated from fermented spider plant or Pak Sian Dong in this study, found ability to present bile salt hydrolase and uricase activity. This isolate is probably able to use for reducing of cholesterol and uric acid in person who is defined as hypercholesterolemia and hyperuricemia. This isolate exhibited resistance to gastrointestinal condition and survived in proper time for using as oral probiotic bacteria. Due to possibility to find the bacteria with bile salt hydrolase and uricase activity in Pak Sian Dong, this kind of fermented food would be established as a health beneficial food and also a good source for probiotic bacteria screening. However, there are several properties of probiotic needs to be conducted in future study for probiotic authentication of the L. brevis SF121 isolate.
ACKNOWLEDGMENTS
We would like to thank Chulabhorn International College of Medicine for all facilities. This research was supported by Thammasat University, Thailand.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human or animal experiments by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in this manuscript.
- Swain MR, Anandharaj M, Ray RC, Rani RP. Fermented fruits and vegetables of Asia: a potential source of probiotics. Biotechnol Res Int. 2014;2014:250424.
Crossref - Azam M, Mohsin M, Ijaz H, et al. Review – Lactic acid bacteria in traditional fermented Asian foods. Pak J Pharm Sci. 2017;30(5):1803-1814. PMID: 29084705
- Barla F, Koyanagi T, Tokuda N, et al. The gamma-aminobutyric acid-producing ability under low pH conditions of lactic acid bacteria isolated from traditional fermented foods of Ishikawa Prefecture, Japan, with a strong ability to produce ACE-inhibitory peptides. Biotechnol Rep (Amst). 2016;10:105-110.
Crossref - Doan NT, Van Hoorde K, Cnockaert M, et al. Validation of MALDI-TOF MS for rapid classification and identification of lactic acid bacteria, with a focus on isolates from traditional fermented foods in Northern Vietnam. Lett Appl Microbiol. 2012;55(4):265-273.
Crossref - Mokoena MP, Mutanda T, Olaniran AO. Perspectives on the probiotic potential of lactic acid bacteria from African traditional fermented foods and beverages. Food Nutr Res. 2016;60:29630.
Crossref - Ouoba LII, Nyanga-Koumou CAG, Parkouda C, et al. Genotypic diversity of lactic acid bacteria isolated from African traditional alkaline-fermented foods. J Appl Microbiol. 2010;108(6):2019-2029.
Crossref - Kumar RS, Kanmani P, Yuvaraj N,Paari KA, Pattukumar V, Arul V. Traditional Indian fermented foods: a rich source of lactic acid bacteria. Int J Food Sci Nutr. 2013;64(4):415-428.
Crossref - Smitinont T, Tansakul C, Tanasupawat S, et al. Exopolysaccharide-producing lactic acid bacteria strains from traditional Thai fermented foods: isolation, identification and exopolysaccharide characterization. Int J Food Microbiol. 1999;51(2-3):105-111.
Crossref - Son SH, Jeon HL, Yang SJ, et al. Probiotic lactic acid bacteria isolated from traditional Korean fermented foods based on beta-glucosidase activity. Food Sci Biotechnol. 2018;27(1):123-129.
Crossref - Yu J, Wang HM, Zha MS, et al. Molecular identification and quantification of lactic acid bacteria in traditional fermented dairy foods of Russia. J Dairy Sci. 2015;98(8):5143-5154.
Crossref - Ji K, Jang NY, Kim YT. Isolation of Lactic Acid Bacteria Showing Antioxidative and Probiotic Activities from Kimchi and Infant Feces. J Microbiol Biotechnol. 2015;25(9):1568-1577.
Crossref - Touret T, Oliveira M, Semedo-Lemsaddek T. Putative probiotic lactic acid bacteria isolated from sauerkraut fermentations. PLoS One. 2018;13(9):e0203501.
Crossref - Yu Z, Zhang X, Li S, Li C, Li D, YangZ. Evaluation of probiotic properties of Lactobacillus plantarum strains isolated from Chinese sauerkraut. World J Microbiol Biotechnol. 2013;29(3):489-498.
Crossref - Onyango CM KC, Ontita EG, Narla RD, Kimenju JW. Current status on production and utilization of spider plant (Cleome gynandra L.) an underutilized leafy vegetable in Kenya. Genet Resour Crop Ev. 2013;60(7):2183-2189.
Crossref - Devi SM, Archer AC, Halami PM. Screening, Characterization and In Vitro Evaluation of Probiotic Properties Among Lactic Acid Bacteria Through Comparative Analysis. Probiotics Antimicrob Proteins. 2015;7(3):181-192.
Crossref - Gotteland M, Cires MJ, Carvallo C, et al. Probiotic screening and safety evaluation of Lactobacillus strains from plants, artisanal goat cheese, human stools, and breast milk. J Med Food. 2014;17(4):487-495.
Crossref - Zhang F, Jiang M, Wan C, et al. Screening probiotic strains for safety: Evaluation of virulence and antimicrobial susceptibility of enterococci from healthy Chinese infants. J Dairy Sci. 2016;99(6):4282-4290.
Crossref - Kumar R, Grover S, Batish VK. Bile Salt Hydrolase (Bsh) Activity Screening of Lactobacilli: In Vitro Selection of Indigenous Lactobacillus Strains with Potential Bile Salt Hydrolysing and Cholesterol-Lowering Ability. Probiotics Antimicrob Proteins. 2012;4(3):162-172.
Crossref - Jones ML, Martoni CJ, Parent M, et al. Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Br J Nutr. 2012;107(10):1505-1513.
Crossref - Wang G, Huang W, Xia Y, et al. Cholesterol-lowering potentials of Lactobacillus strain overexpression of bile salt hydrolase on high cholesterol diet-induced hypercholesterolemic mice. Food Funct. 2019;10(3):1684-1695.
Crossref - Kelley WN, Wyngaarden JB. Drug treatment of gout. Semin Drug Treat. 1971;1(2):119-147.
- Biggers K, Scheinfeld N. Pegloticase, a polyethylene glycol conjugate of uricase for the potential intravenous treatment of gout. Curr Opin Investig Drugs. 2008;9(4):422-429.
- Nanda P, Babu PE. Isolation, screening and production studies of uricase producing bacteria from poultry sources. Prep Biochem Biotechnol. 2014;44(8):811-821.
Crossref - Mahler JL. A new bacterial uricase for uric acid determination. Anal Biochem. 1970;38(1):65-84.
Crossref - Shaaban MI, Abdelmegeed E, Ali YM. Cloning, expression, and purification of recombinant uricase enzyme from Pseudomonas aeruginosa Ps43 using Escherichia coli. J Microbiol Biotechnol. 2015;25(6):887-892.
Crossref - Iswantini D, Nurhidayat N, Trivadila, et al. Activity and stability of uricase from Lactobacillus plantarum immobilizated on natural zeolite for uric acid biosensor. Pak J Biol Sci. 2014;17(2):277-281.
Crossref - Yasiri A, Vannaxay, E, Kiatmontri J,d Seubsasana S. Isolation and determination of bile salt hydrolase-producing lactic acid bacteria from fermented spider plant. J Pure Appl Microbiol. 2018;12(3):1055-1060.
Crossref - Mota RM, Moreira JL, Souza MR, et al. Genetic transformation of novel isolates of chicken Lactobacillus bearing probiotic features for expression of heterologous proteins: a tool to develop live oral vaccines. BMC Biotechnol. 2006;6:2.
Crossref - Tanasupawat S, Komagata K. Lactic acid bacteria in fermented foods in Thailand. World J Microbiol Biotechnol. 1995;11(3):253-256.
Crossref - Zhang B, Wang Y, Tan Z, et al. Screening of probiotic activities of lactobacilli strains isolated from traditional Tibetan Qula, a raw yak milk cheese. Asian-Australas J Anim Sci. 2016;29(10):1490-1499.
Crossref - Tuo Y, Yu H, Ai L, et al. Aggregation and adhesion properties of 22 Lactobacillus strains. J Dairy Sci. 2013;96(7):4252-7.
Crossref - Ronka E, Malinen E, Saarela M, et al. Probiotic and milk technological properties of Lactobacillus brevis. Int J Food Microbiol. 2003;83(1):63-74.
Crossref - Shakibaie M, Mohammadi-Khorsand T, Adeli-Sardou M, et al. Probiotic and antioxidant properties of selenium-enriched Lactobacillus brevis LSe isolated from an Iranian traditional dairy product. J Trace Elem Med Biol. 2017;40:1-9.
Crossref - Fang F, Xu J, Li Q, et al. Characterization of a Lactobacillus brevis strain with potential oral probiotic properties. BMC Microbiol. 2018;18(1):221.
Crossref - Song MW, Jang HJ, Kim KT, et al. Probiotic and Antioxidant Properties of Novel Lactobacillus brevis KCCM 12203P Isolated from Kimchi and Evaluation of Immune-Stimulating Activities of Its Heat-Killed Cells in RAW 264.7 Cells. J Microbiol Biotechnol. 2019;29(12):1894-903.
Crossref - Kariyawasam K, Yang SJ, Lee NK, et al. Probiotic Properties of Lactobacillus brevis KU200019 and Synergistic Activity with Fructooligosaccharides in Antagonistic Activity against Foodborne Pathogens. Food Sci Anim Resour. 2020;40(2):297-310.
Crossref - Gu XC, Luo XG, Wang CX, et al. Cloning and analysis of bile salt hydrolase genes from Lactobacillus plantarum CGMCC No. 8198. Biotechnol Lett. 2014;36(5):975-983.
Crossref - Lambert JM, Bongers RS, de Vos WM, et al. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl Environ Microbiol. 2008;74(15):4719-4726.
Crossref - Kumar R, Grover S, Batish VK. Hypocholesterolaemic effect of dietary inclusion of two putative probiotic bile salt hydrolase-producing Lactobacillus plantarum strains in Sprague-Dawley rats. Br J Nutr. 2011;105(4):561-573.
Crossref - Rani RP, Anandharaj M, Ravindran AD. Characterization of Bile Salt Hydrolase from Lactobacillus gasseri FR4 and Demonstration of Its Substrate Specificity and Inhibitory Mechanism Using Molecular Docking Analysis. Front Microbiol. 2017;8:1004.
Crossref - Travers MA, Sow C, Zirah S, et al. Deconjugated bile salts produced by extracellular bile-salt hydrolase-like activities from the probiotic Lactobacillus johnsonii La1 inhibit Giardia duodenalis in vitro growth. Front Microbiol. 2016;7:1453.
Crossref - Elkins CA, Savage DC. Identification of genes encoding conjugated bile salt hydrolase and transport in Lactobacillus johnsonii 100-100. J Bacteriol. 1998;180(17):4344-4349.
Crossref - Reyes-Nava L, Garduno-Siciliano L, Estrada-de los Santos P, et al. Use of bile acids as a selection strategy for lactobacillus strains with probiotic potential. J Food Nutr Disor. 2016;5(1).
- Huang Y, Wang X, Wang J, et al. Lactobacillus plantarum strains as potential probiotic cultures with cholesterol-lowering activity. J Dairy Sci. 2013;96(5):2746-2753.
Crossref - Alvarez-Lario B, Macarron-Vicente J. Uric acid and evolution. Rheumatology (Oxford). 2010;49(11):2010-2015.
Crossref - Chen RJ, Chen MH, Chen YL, et al. Evaluating the urate-lowering effects of different microbial fermented extracts in hyperuricemic models accompanied with a safety study. J Food Drug Anal. 2017;25(3):597-606.
Crossref - Szczurek P, Mosiichuk N, Wolinski J, et al. Oral uricase eliminates blood uric acid in the hyperuricemic pig model. PLoS One. 2017;12(6):e0179195.
Crossref - Yu Y, Liu Q, Li H, Wen C, He Z. Alterations of the Gut Microbiome Associated With the Treatment of Hyperuricaemia in Male Rats. Front Microbiol. 2018;9:2233.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.