ISSN: 0973-7510
E-ISSN: 2581-690X
Small ruminants, particularly goats are susceptible to respiratory diseases. Respiratory diseases, especially pneumonia and associated mortality causes significant threat to the goat farming due to substantial economic losses in the eastern plain zone of Uttar Pradesh, India. Among the respiratory infections in goats, Pasteurella spp., Escherichia coli (E. coli), Klebsiella spp. and Staphylococcus spp. infections in eastern plain zone of Uttar Pradesh had been studied limited. To determine the occurrence of respiratory diseases among goats, the current study was designed with an objective of isolation, identification and molecular characterization of organisms responsible for respiratory infections, especially Pasteurella spp., E. coli, Klebsiella spp. and Staphylococcus spp. A total of 150 samples were collected during postmortem examination of goats and processed for isolation of respiratory pathogens, genomic characterization, pathological lesions, localization of organisms in tissues by immunohistochemistry, and scanning electron microscopy (SEM). Out of 150 samples, 40.66% of E. coli, 26% Pasteurella spp., 20% Klebsiella spp., and 13.33% Staphylococcus spp. were identified in the respiratory tract of goats based on their typical morphology, colony characteristics, bio-chemical properties, and motility. The samples crucial for diagnosis of respiratory bacterial pathogens were lungs, followed by heart blood, nasal swabs, and tracheal swabs. Gross pathological lesions were consolidation in cranio-ventral and cranio-lateral lobes of lungs, congestion, hemorrhages, and fibrinous pleuritis. Histopathological lesions were bronchopneumonia with infiltration of neutrophils, hemorrhages, and necrotic regions contained spindle-shaped/elongated “oat cells or streaming leukocytes”. Loss of cilia was noticed by SEM. In lungs and intestine, Pasteurella spp. and E. coli antigen were localized in the cytoplasm of neutrophils, alveolar macrophages, bronchiolar epithelial cells, and intestinal mucosa. Extraintestinal pathogenic E. coli was found in the dead goats with pneumonia. Most of the isolated Pasteurella strains were having similarities with Pasteurella multocida based on the phylogenetic analysis targeting 16S rRNA gene. Results of the present study confirmed the circulation of Pasteurella spp., E. coli, Klebsiella spp. and Staphylococcus spp. among goats in field conditions in eastern plain zone of Uttar Pradesh and necessary control measures should be formulated with effective vaccination strategies in small ruminants for the control of respiratory infections to reduce the economic losses to goat farmers.
Goats, Respiratory Infections, Pasteurella multocida, Escherichia coli, Phylogenetic Analysis, Isolation, Pathology
India has a rapidly expanding animal husbandry industry that accounts for 11.6% (528 million) of the world’s livestock population and working towards becoming self-sufficient in the production of livestock products.1,2 Among farm animals, goats are considered as “poor farmer’s mobile bank” and India is second in the world in terms of population of goats (148.88 million, as per the 20th Livestock Census-2019). The economy of farmers in the arid, semiarid, and hilly areas of India are greatly influenced by goats. Goats are the most valuable asset due to their rapid growth rate as well as their considerable contribution to the meat, milk, and fiber production.3
Goats are highly vulnerable to respiratory ailments, which account for roughly 50% of their fatalities.4 Respiratory diseases due to infectious agents in sheep and goats account for 5.6% of all small ruminant illnesses, regardless of their underlying causes.2 According to Lacasta et al.,5 the respiratory disease complex in small ruminants is caused by several different etiological factors. Due to their significant clinical manifestations, bacterial diseases have drawn particular attention.4
Both humans and animals can get a range of intestinal and extra-intestinal diseases from pathogenic Escherichia coli (E. coli) and Pasteurella spp.6 Analogous to intestinal diseases, respiratory diseases pose a significant risk to the goat breeding sector.7 Gram-negative bacterium Pasteurella multocida (P. multocida) is pathogenic and causes pasteurellosis. P. multocida is categorized into five capsular serogroups, 3 subspecies and 16 serotypes.8 Worldwide, in ruminants of all ages, P. multocida causes bronchopneumonia, which has a cranio-ventral lung distribution. It is an opportunistic pathogen and frequently reside in the upper respiratory tract as commensals, but they can also invade the tissues of animals with impaired immune system.2 According to Chakraborty et al.,9 animals with compromised immune system or under stress such as pregnancy, nursing, young, or aged animals are more susceptible to respiratory pathogens like P. multocida, Mannheimia haemolytica (formerly Pasteurella haemolytica), and other species.
Klebsiella spp. is a Gram-negative, rod-shaped, and capsulated bacterium belongs to the family Enterobacteriaceae. Recently, Klebsiella pneumoniae (K. pneumoniae) is receiving increased attention worldwide due to severe disease, biofilm formation, and antibiotic resistance.10,11 K. pneumoniae is a virulent bacterium and significant cause of pneumonia in sheep and goats. Clinical signs are pyrexia, loss of appetite, nasal discharge, difficulty breathing, rapidly progressing course, and complication of lung abscess with involvement of multiple lobes. K. pneumoniae can also be transmitted to humans from goats.10-12 Staphylococcus spp. normally found in the upper respiratory tract of healthy sheep and goat; however, during stress conditions like overcrowding, inadequate ventilation, poor nutrition and transportation, or in association with other pathogens causes pneumonia.13,14 Clinical signs are pyrexia, sudden death without warning signs, and increased respiratory rate with abdominal efforts. Staphylococcus aureus (S. aureus) can be resistant to antibiotics and can be transmitted between animals and humans.13-15
Pneumonic animals exhibit a loss of appetite or anorexia, lethargy, coughing, fever, dyspnea, and mucopurulent nasal discharge.2,4 Young animals are more susceptible to infection compared to adults, which might result in rapid death with or without any prior clinical indications.12,15 Animals and birds that died of respiratory diseases due to bacterial infections exhibit ventral consolidation in the cranial lobes of the lungs, as well as fibrinous pericardial and pleural effusions.12,15,16 The most accurate way to diagnose respiratory infections is to isolate the bacteria from the lungs and nasopharyngeal swabs of suspicious animals, followed by biochemical confirmation of the isolates. Using bacteriological techniques, it is frequently difficult to identify Pasteurella spp., E. coli, K. pneumoniae, and S. aureus during various circumstances, including antibiotic therapy, frozen tissue, autolytic material, and other situations. However, immunohistochemistry (IHC) technique, overcomes some difficulties with earlier methods of detecting Pasteurella antigens on formalin-fixed and paraffin-embedded tissues.17,18 Molecular methods like polymerase chain reaction (PCR) have recently been found to be incredibly sensitive and specific.19 Caprine pasteurellosis, E. coli, K. pneumoniae, and S. aureus infections have been given less attention and only a small number of investigations have been conducted. The current study was undertaken to determine the occurrence of the respiratory diseases with isolation, identification and molecular characterization of the causative organisms especially Pasteurella spp., E. coli, K. pneumoniae, and S. aureus among goats of Eastern Uttar Pradesh. The results of the current study could serve as useful information for the concerned authorities in designing the proper therapeutic and control measures and to reduce the economic losses of goat farmers.
Sample collection
The samples for the present study were collected from the natural cases of respiratory infections in goats from September 2021 to August 2022. Postmortem examinations of 150 goat carcasses suspected of respiratory diseases were conducted. Among 150 goats, 36 were from Azamgarh, 40 from Ghazipur, 24 from Mau, 34 from Ballia, 14 from Bhadohi, and 2 from Varanasi districts of the eastern plain zone of Uttar Pradesh. A total of 51 samples were collected from ≤6 months, 39 samples from >6 months to ≤1 year, 32 samples from >1 year to ≤2 years, 17 samples from >2 year to ≤3 years, and 11 samples from >3 years. Samples were collected from 109 female and 41 male goats. Further, 76 samples were collected from non-descript goats, 43 samples from Jamunapari goats and 31 samples from Barbari goats. The died goats were not vaccinated and dewormed against any diseases. Among 150 goats, 67 samples were collected during winter (mid-November to February), 28 samples during summer (March to mid-June), 36 samples during monsoon (mid-June to September), and 19 samples during post-monsoon (October and mid-November). The died goats were from unorganized farm, extensive system of rearing with free grazing in pasture lands and tree foliage, and housed in open housing system or thatched roof with wooden/bamboo sticks with an earthen floor. The goats belonged to the landless, small and marginal farmers had the flock size between 5 to 25. Heart blood, nasal and tracheal swabs, and lungs were collected in the sterile container for bacterial isolation. The tissue samples were collected from the cases showing gross lesions in the lungs, trachea, intestine, and lymph nodes in 10% neutral buffered formalin (NBF; Cat No. F1635-4L, Sigma-Aldrich, St. Louis, Missouri, USA) for histopathology and immunohistochemistry. Lungs were collected in 2.5% glutaraldehyde (Cat No. G5882-100ML, Sigma-Aldrich, St. Louis, Missouri, USA) for electron microscopy.
Isolation of Escherichia coli
A total of 150 nasal swabs and lung tissues from dead goats were processed for bacterial isolation. A loop of inoculum from the nutrient broth (Cat No. M002-500G, HiMedia Laboratories Private Limited, Thane, Maharashtra, India) was streaked onto MacConkey agar (MCA; Cat No. MH081-500G, HiMedia Laboratories Private Limited, Thane, Maharashtra, India) plates to isolate the lactose-fermenting and non-lactose-fermenting bacteria. A bright red or pink color colony was used to culture in eosin methylene blue (EMB) agar (Cat No. M317-500G, HiMedia Laboratories Private Limited, Thane, Maharashtra, India). The colonies of red or pink and greenish black in color with metallic sheen were considered as positive for Escherichia coli in EMB agar. The positive colonies were subculture into EMB agar to obtain pure colonies. Again, the pure colonies were subjected to sugar fermentation and other biochemical tests namely indole, catalase, Voges-Proskauer (VP), methyl red (MR), citrate utilization, nitrate reduction, triple sugar iron (TSI) agar, hydrogen sulfide (H2S) production, and urease production tests as per the procedures outlined by Quinn et al.20
Isolation of Pasteurella multocida
Blood agar (BA; Cat No. M1301-500G, HiMedia Laboratories Private Limited, Thane, Maharashtra, India) and MacConkey agar were used as primary culture media for the initial isolation of organisms from the samples as per the procedures outlined by Quinn et al.20 Initial speculation suggested that Pasteurella multocida was present in the isolates that failed to grow on MCA. A single non-haemolytic colony of these isolates was selected from the primary culture and re-streaked on a fresh BA plate. The cultures were stained with Gram’s staining (Cat No. K001-1KT, HiMedia Laboratories Private Limited, Thane, Maharashtra, India) to determine whether the growth and morphology of the organisms were pure. The cultures were inoculated into the nutrient agar slant and stored at 4 °C for further identification. Identification of P. multocida was done based on growth morphology and biochemical characteristics as per the method described by Edward and Ewing.21
Isolation of Klebsiella spp. and Staphylococcus spp.
A loop of inoculum from the nutrient broth was streaked onto MacConkey agar plates to isolate the lactose-fermenting and non-lactose-fermenting bacteria. A lactose-fermenting, large, mucoid, and red colony was used to culture in EMB agar to isolate the Klebsiella spp. The mannitol salt agar (Cat No. MH118-500G, HiMedia Laboratories Private Limited, Thane, Maharashtra, India) plates were streaked with a loopful of inoculum from nutrient broth to isolate the Staphylococcus aureus as per the procedures outlined by Quinn et al.20
Demonstration of bipolar organisms
The smears of the colonies from blood agar were prepared and stained with Giemsa stain (Cat No. TC232-5G, HiMedia Laboratories Private Limited, Thane, Maharashtra, India) for demonstration of bipolar organisms.
Isolation of bacterial genomic DNA
Genomic DNA was isolated from the bacterial samples after biochemical characterization. The samples were homogenized with extraction buffer (Cat No. 56304, QIAamp® DNA Micro Kit, Qiagen, Maryland, USA). Equal volume of phenol, chloroform, and isoamyl alcohol (25:24:1, respectively) were added and mixed well. The tubes were centrifuged at 14,000 rpm for 15 min at room temperature (RT). After collecting the upper aqueous phase in a fresh tube, the mixture of equal parts of chloroform:isoamyl alcohol (24:1) was added. After centrifuging for 10 min at 14,000 rpm in RT, the upper aqueous phase was collected and transferred into a fresh tube. By adding 100 µL of 3 M sodium acetate, pH 7.0 and 700 µL of isopropanol to the mixture, the DNA was isolated and precipitated. The tubes were incubated for 15 min at RT, and centrifuged for 15 min at 14,000 rpm on 4 °C. After twice washing, the DNA pellet in 70% ethanol and quickly rinsed again in 100% ethanol and allowed to air dry. In Tris-EDTA (TE) buffer (Tris-Cl, 10 mM, pH 8.0; EDTA, 1 mM), the DNA was dissolved. In order to remove the RNA, 5 µl of DNAse free RNAse (10 mg/mL, Cat No. FEREN0531, Thermo Fisher Scientific, Carlsbad, California, USA) was added to the DNA as per the procedures described by Ingale et al.22
PCR amplification of 16S rRNA gene
Isolated bacterial genomic DNA was amplified using universal bacterial 16S ribosomal RNA (16S rRNA) gene primers 27F (5’-AGA GTT TGA TCC TGG CTC AG-3’) and 1392R (5’-GGT TAC CTT GTT ACG ACT T-3’),23 which covers nearly full length of 16S rRNA gene with expected amplicon of >1,350 bp. Polymerase chain reaction (PCR) was performed to amplify the 16S rRNA gene using 10 ng of genomic DNA extracted from each bacterial isolate and 10 pM concentrations of each forward and reverse primers as per the procedures described by Srinivasan et al.23 The amplified PCR products were visualized on a 1% agarose gel stained with ethidium bromide under UV light to confirm the presence of a >1,350 bp band. Pasteurella multocida specific PCR (PM-PCR) was performed using published primers24 and protocols described by Townsend et al.24 The serotype of the E. coli isolates was identified by O-genotyping PCR using published primers25 and protocols described by Iguchi et al.25
Sequencing of bacterial isolates
The PCR products were sequenced Bi-directionally.26 The amplified products of 16S rRNA by PCR with universal primer 27F-1392R were purified and submitted for sequencing.
Phylogenetic analysis
The sequence data was aligned and analyzed to identify the bacterial species and its closest neighbours. The obtained forward and reverse sequences were aligned using the online pairwise alignment tool BioEdit.27 The query sequences were identified by considering E-value 1 × 10-5 and maximum hits (99% or 100%) with a species in the reference database National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/). In addition to Basic Local Alignment Search Tool (BLAST; https://blast.ncbi.nlm.nih.gov/), Molecular Evolutionary Genetics Analysis Version 7.0 (MEGA7) software was used for phylogenetic tree analysis to conduct the evolutionary analyses28 by employing Maximum Likelihood (ML) method.26
Necropsy examination
Systematic necropsy examinations of 150 goat carcasses were carried. The gross pathological lesions in different visceral organs were recorded.
Histopathology
After proper fixation in 10% NBF, tissue samples from the lungs, trachea, intestine, and lymph nodes were processed for routine paraffin wax embedding. Tissue sections of 4-5 µ thickness were cut and stained with hematoxylin and eosin (H&E) method for histopathological examination as per the standard procedure.29
Immunohistochemistry
Tissue sections were taken on the (3-aminopropyl)triethoxysilane (APES; Cat No. 440140-100ML, Sigma-Aldrich, St. Louis, Missouri, USA) coated glass slides to carry out IHC. Immunohistochemical staining was done using polyclonal antibodies as per the method described by Saminathan et al.30 Further, the intensity of the IHC reaction was determined based on the antigen-positive cells and graded as mild, moderate, or marked. Scoring was done by the panel of two Pathologists. Tissue sections were examined in low power (10X objective lens) and high power (40X objective lens) fields and the entire tissue sections were examined for scoring purposes. If no staining, it was considered as negative, if 5 to 10% stained cells as mild, if more than 10 to 25% stained cells as moderate, and if more than 25% stained cells as marked IHC reaction.
Special staining
Special staining such as Leishman’s staining (Cat No. GRM949-25G, HiMedia Laboratories Private Limited, Thane, Maharashtra, India) for Pasteurella bipolar organisms, Von Kossa stain for the demonstration of calcium, and Masson’s trichrome stain for connective tissue fibers were done as per the standard procedure.29
Scanning electron microscopy
For scanning electron microscopy, specimens were fixed with 3.5 N hydrochloric acid (HCl) for 3 weeks at RT. Specimens were treated with 0.5% tannic acid solution. Post-fixation was done by immersing in 1% osmium tetroxide (OsO4) solution for 10 min. Specimens were dehydrated with a graded series of ethanol (70-100%). Dehydrated specimens were freeze-dried with a t-butyl alcohol freeze-drying method. Specimens were coated with platinum (Pt)-palladium (Pd) and observed under scanning electron microscope (S-800, Hitachi-Hi-Technologies, Japan).
Statistical analysis
Statistical Package for the Social Sciences (SPSS) software (version 17.0) was used to perform the standard statistical procedures on the data obtained on various parameters. The chi-square test was utilized according to the WEB AGRI STAT PACKAGE programme, which was created at the ICAR Research Centre, Goa, India.31
Clinical signs
Goats affected with respiratory infections exhibited pyrexia with a temperature of
40-41 °C, anorexia, wet cough with crackling sounds, dyspnea, serous to mucopurulent nasal and ocular discharges, increased respiration, abdominal breathing, and sudden death without clinical signs.
Isolation and identification of bacteria from respiratory infections
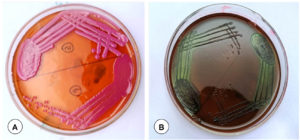
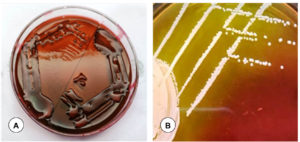
Among 150 samples, based on the typical colony characteristics (motility, morphology, and bio-chemical properties), 61 (40.66%) samples were identified as positive for E. coli, and 39 (26%) samples for Pasteurella spp., 30 (20%) samples for Klebsiella spp., and 20 (13.33%) samples for Staphylococcus spp. in the respiratory tract of goats. A total of 96 lung samples, 84 heart blood, 46 nasal swabs, and 29 tracheal swabs from the goats that were clinically suspected and later died of respiratory infections were showed positivity for the respiratory pathogens by bacteriological screening. On MacConkey agar, E. coli isolates produced big (2-3 mm) and pink colonies that were fermented lactose (Figure 1A). On EMB agar, E. coli produced the distinctive “metallic shine” colonies (Figure 1B). The rose-pink colonies with a mucoid appearance and also colonies of light purple mucoid appearance on EMB agar were construed as Klebsiella spp. (Figure 2A). Staphylococcus spp. colonies on solid media (Mannitol salt agar) were round, smooth, and glistening with shining surface (Figure 2B). No growth on MacConkey agar is indicative of Pasteurella spp. (Figure 3A).
Figure 1. (A). Growth of E. coli on MacConkey lactose agar visible as lactose-fermenting pink color colonies. (B). Growth of E. coli. on eosin methylene blue (EMB) agar visible as blue-black with a dark center and metallic sheen
Figure 2. (A). Dark purple mucoid colonies on eosin methylene blue (EMB) agar indicating Klebsiella spp. (B). Yellow colour in the region of medium where the Staphylococcus grown due to mannitol fermentation on mannitol salt agar
Figure 3. (A). No growth on MacConkey lactose agar indicating Pasteurella spp. (B). Pasteurella spp. appeared as rod-shaped with bipolar appearance (hairpin-like shape) in tissue impression smears by Leishman’s staining
Morphology of bacteria
The isolated non-hemolytic and hemolytic individual colonies were cultured on a blood agar plate, and the smears of colonies from the blood agar plate were stained with Gram stain. A total of 130 isolates were Gram-negative and 20 isolates were Gram-positive by Gram staining. Pasteurella spp. showed coccobacilli by Gram staining and in Leishman’s staining, Pasteurella spp. appeared as rod-shaped with bipolar appearance (hairpin-like shape), because the ends of the bacteria stained more intensely than the middle (Figure 3B). E. coli and Klebsiella spp. showed rod shaped, and Staphylococcus spp. showed cocci in clusters, pairs and occasionally in short chains of organisms by Gram staining.
Biochemical characterization of Pasteurella spp.
All 39 isolates of Pasteurella spp. were subjected to biochemical tests. The percent positivity for Pasteurella multocida was 74.35% (29); while, the percent positivity of Mannheimia haemolytica was 25.64% (10). The results of biochemical tests were shown in Table 1.
Table (1):
Biochemical test results of Pasteurella multocida and Mannheimia haemolytica
| Biochemical test | Species of Pasteurella isolated/suspected | |
|---|---|---|
| P. multocida (29) | Mannheimia haemolytica (10) | |
| Catalase | ++ | + |
| Oxidase | + | + |
| Urease | – | – |
| Voges-Proskauer (VP) | + | – |
| Methyl red | + | – |
| Sucrose | + | + |
| Indole | – | + |
| Hemolysis | – | + |
| Citrate | – | + |
Biochemical characterization of Escherichia coli
All the E. coli isolates showed typical biochemical reactions that were positive to the indole, MR, TSI agar, and nitrate reduction tests; while, negative to the VP and H2S production tests. E. coli isolates were negative for the citrate utilization and urease activity tests. All the E. coli isolates were motile and isolates fermented D-glucose, lactose, and mannitol.
Molecular confirmation of bacterial isolates
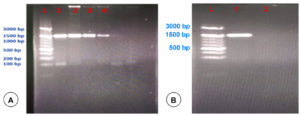
Genomic DNA of Pasteurella spp. (39) and E. coli (61) isolates identified by bacteriological methods were used for amplification and confirmation using PCR by targeting the 16S rRNA gene. All the tested Pasteurella spp. (Figure 4A) and E. coli (Figure 4B) isolates yielded positive results. Out of 39, 31 isolates yielded positive results by Pasteurella multocida specific PCR (PM-PCR). Out of 61 E. coli isolates, 52 were identified as O serotype by O-genotyping PCR.
Figure 4. PCR amplification of 16S rRNA gene of Pasteurella spp. (A) and E. coli (B). Lane L: 100 bp DNA ladder and L1-L4: Samples
Nucleotide sequence and phylogenetic analysis
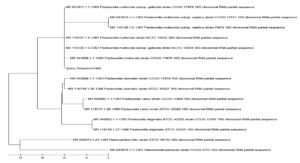
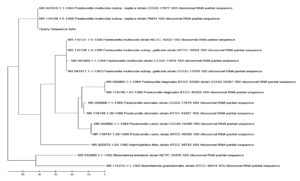
Sequencing of 16S rRNA gene of Pasteurella spp. and E. coli isolates, which were confirmed as P. multocida and E. coli by specific PCR were done. The P. multocida isolates were showed more than 99% similarity with those P. multocida strains submitted in GenBank (accession numbers: NR_041809.1, NR_041811.1, NR_041810.1, and NR_115138.1), according to the results of BLAST analysis. The sequences of some isolates showed highest identity (99.69%) with those of the 16S rRNA of P. multocida strain CCUG 17976 (Figure 5). The sequences of some isolates showed close similarity (98%) with the P. multocida subsp. gallicida strain CCUG 17978 (Figure 6). The sequences of some isolates showed close similarity (99.05%) with the P. multocida subsp. septica strain PM24 (accession no. NR_115138.1) and strain CCUG 17977 (Figure 7). The sequences of E. coli isolates were showed similarity with the E. coli (Figure 8).
Figure 5. Phylogenetic analysis of 16S rRNA gene of Pasteurella spp. positive sample from goat showed highest similarity (99.69%) with P. multocida strain CCUG 17976 16S rRNA
Figure 6. Phylogenetic analysis of 16S rRNA gene of Pasteurella spp. positive sample from goat showed highest similarity (98.83%) with P. multocida subsp. gallicida strain CCUG 17978 16S rRNA
Figure 7. Phylogenetic analysis of 16S rRNA gene of Pasteurella spp. positive sample from goat showed highest similarity (99.05%) with P. multocida subsp. septica strain CCUG 17977 16S rRNA
Figure 8. Phylogenetic analysis of 16S rRNA gene of Escherichia coli positive sample from goat showed highest percentage of identity with E. coli
Occurrence of respiratory pathogens in eastern plain zone of Uttar Pradesh
The overall occurrence of Pasteurella spp. and pathogenic E. coli in the goats of eastern plain zone of Uttar Pradesh was found to be 26% (39 samples) and 40.66% (61 samples), respectively. District-wise occurrence of Pasteurella spp. was found to be 27.77%, 32.5%, 20.58%, 33.33%, 0%, and 50% in the districts of Azamgarh, Ghazipur, Ballia, Mau, Bhadohi, and Varanasi, respectively. Occurrence of Klebsiella spp. and Staphylococcus spp. in the goats of eastern plain zone of Uttar Pradesh were presented in the Table 2. The Klebsiella spp. showed significant difference (p < 0.01) in the occurrence of respiratory pathogens between the various districts of the eastern plain zone of Uttar Pradesh, India (Table 2 and 3).
Table (2):
Occurrence of respiratory pathogens among different districts of eastern plain zone of Uttar Pradesh
| Districts | Number of samples tested | Pasteurella spp. | Escherichia coli | Klebsiella spp. | Staphylococcus spp. | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number of positive | Percent positivity | Number of positive | Percent positivity | Number of positive | Percent positivity | Number of positive | Percent positivity | ||
| Azamgarh | 36 | 10 | 27.77 | 16 | 44.44 | 8 | 22.22 | 2 | 5.55 |
| Ghazipur | 40 | 13 | 32.50 | 20 | 50.00 | 2 | 5.00 | 5 | 12.50 |
| Ballia | 34 | 7 | 20.58 | 13 | 38.23 | 10 | 29.41 | 4 | 11.76 |
| Mau | 24 | 8 | 33.33 | 10 | 41.66 | 2 | 8.33 | 4 | 16.66 |
| Bhadohi | 14 | 0 | 0 | 1 | 7.14 | 8 | 57.14 | 5 | 35.71 |
| Varanasi | 2 | 1 | 50.00 | 1 | 50.00 | 0 | 0 | 0 | 0 |
| Total | 150 | 39 | 26.00 | 61 | 40.67 | 30 | 20.00 | 20 | 13.33 |
Table (3):
District-wise comparison of respiratory pathogens by chi-square test
| Parameters | Pasteurella spp. | Escherichia coli | Klebsiella spp. | Staphylococcus spp. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | df | Asymptotic significance (2-sided) | Value | df | Asymptotic significance (2-sided) | Value | df | Asymptotic significance (2-sided) | Value | df | Asymptotic significance (2-sided) | |
| Pearson chi-square | 2.087a | 4 | 0.720 | 8.343b | 5 | 0.138 | 21.508c | 4 | 0.000 | 8.183d | 4 | 0.085 |
| Likelihood ratio | 2.103 | 4 | 0.717 | 9.961 | 5 | 0.076 | 21.118 | 4 | 0.000 | 7.129 | 4 | 0.129 |
| Linear-by-linear association | 0.035 | 1 | 0.852 | 3.547 | 1 | 0.060 | 3.793 | 1 | 0.051 | 6.092 | 1 | 0.014 |
| N of valid cases | 136 | 150 | 148 | 148 | ||||||||
Note: df – Degrees of freedom; Superscript ‘a’ indicate 2 cells (20.0%) have expected count less than 5. The minimum expected count was 0.57; ‘b’ indicates 2 cells (16.7%) have expected count less than 5. The minimum expected count was 0.81; ‘c’ indicates 2 cells (20.0%) have expected count less than 5. The minimum expected count was 2.84; ‘d’ indicates 4 cells (40.0%) have expected count less than 5. The minimum expected count was 1.89. Except Klebsiella spp. (p<0.01), there was no significant difference among districts regarding respiratory pathogens.
Sex-, age-, breed-, and season-wise occurrence of respiratory pathogens
The overall sex-, age-, breed-, and season-wise occurrence of Pasteurella spp.,
E. coli, Klebsiella spp. and Staphylococcus spp. in the goats of eastern plain zone of Uttar Pradesh were presented in the Table 4-11.
Table (4):
Occurrence of respiratory pathogens among different sex in eastern plain zone of Uttar Pradesh
| Sex | Number of samples tested | Pasteurella spp. | Escherichia coli | Klebsiella spp. | Staphylococcus spp. | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number of positive | Percent positivity | Number of positive | Percent positivity | Number of positive | Percent positivity | Number of positive | Percent positivity | ||
| Male | 41 | 9 | 21.95 | 22 | 53.65 | 6 | 14.63 | 4 | 9.76 |
| Female | 109 | 30 | 27.52 | 39 | 35.77 | 24 | 22.02 | 16 | 10.67 |
| Total | 150 | 39 | 26.00 | 61 | 40.67 | 30 | 20.00 | 20 | 13.33 |
Table (5):
Sex-wise comparison of respiratory pathogens by chi-square test
| Parameters | Pasteurella spp. | Escherichia coli | Klebsiella spp. | Staphylococcus spp. | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | df | Asymptotic significance (2-sided) | Exact significance (2-sided) | Exact significance (1-sided) | Value | df | Asymptotic significance (2-sided) | Exact significance (2-sided) | Exact significance (1-sided) | Value | df | Asymptotic significance (2-sided) | Exact significance (2-sided) | Exact significance (1-sided) | Value | df | Asymptotic significance (2-sided) | Exact significance (2-sided) | Exact significance (1-sided) | |
| Pearson chi-square | 0.481a | 1 | 0.488 | 3.947b | 1 | 0.047 | 1.015c | 1 | 0.314 | 0.625d | 1 | 0.429 | ||||||||
| Continuity correctione | 0.235 | 1 | 0.628 | 3.241 | 1 | 0.072 | .606 | 1 | 0.436 | 0.271 | 1 | 0.602 | ||||||||
| Likelihood ratio | 0.492 | 1 | 0.483 | 3.902 | 1 | 0.048 | 1.067 | 1 | 0.302 | 0.660 | 1 | 0.416 | ||||||||
| Fisher’s exact test | 0.538 | 0.319 | 0.062 | 0.036 | 0.367 | 0.221 | 0.592 | 0.310 | ||||||||||||
| Linear-by-linear association | 0.478 | 1 | 0.490 | 3.921 | 1 | 0.048 | 1.009 | 1 | 0.315 | 0.621 | 1 | 0.431 | ||||||||
| N of valid cases | 150 | 150 | 150 | |||||||||||||||||
Note: df – Degrees of freedom; Superscript ‘a’ indicate 0 cells (0.0%) have expected count less than 5. The minimum expected count was 10.66; ‘b’ indicates 0 cells (0.0%) have expected count less than 5. The minimum expected count was 16.67; ‘c’ indicates 0 cells (0.0%) have expected count less than 5. The minimum expected count was 8.20; ‘d’ indicates 0 cells (0.0%) have expected count less than 5. The minimum expected count was 5.47; ‘e’ indicates computed only for a 2×2 table. Except Escherichia coli (p<0.05), there was no significant difference among male and female regarding respiratory pathogens.
Table (6):
Occurrence of respiratory pathogens among different age groups in eastern plain zone of Uttar Pradesh
| Age groups | Number of samples tested | Pasteurella spp. | Escherichia coli | Klebsiella spp. | Staphylococcus spp. | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number of positive | Percent positivity | Number of positive | Percent positivity | Number of positive | Percent positivity | Number of positive | Percent positivity | ||
| ≤ 6 months | 51 | 15 | 29.41 | 25 | 49.01 | 6 | 11.76 | 5 | 9.80 |
| > 6 months to ≤ 1 year | 39 | 10 | 25.64 | 16 | 41.02 | 8 | 20.51 | 5 | 12.82 |
| > 1 year to ≤ 2 years | 32 | 8 | 25.00 | 11 | 34.37 | 9 | 28.12 | 4 | 12.50 |
| > 2 year to ≤ 3 years | 17 | 4 | 23.52 | 6 | 35.29 | 3 | 17.64 | 4 | 23.52 |
| > 3 years | 11 | 2 | 18.18 | 3 | 27.27 | 4 | 36.36 | 2 | 18.18 |
| Total | 150 | 39 | 26.00 | 61 | 40.67 | 30 | 20.00 | 20 | 13.33 |
Table (7):
Age-wise comparison of respiratory pathogens by chi-square test
| Parameters | Pasteurella spp. | Escherichia coli | Klebsiella spp. | Staphylococcus spp. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | df | Asymptotic Significance (2-sided) | Value | df | Asymptotic Significance (2-sided) | Value | df | Asymptotic Significance (2-sided) | Value | df | Asymptotic Significance (2-sided) | |
| Pearson chi-square | 0.731a | 4 | 0.947 | 3.023b | 4 | 0.554 | 5.388c | 4 | 0.250 | 2.331d | 4 | 0.675 |
| Likelihood ratio | 0.752 | 4 | 0.945 | 3.054 | 4 | 0.549 | 5.307 | 4 | 0.257 | 2.120 | 4 | 0.714 |
| Linear-by-linear association | 0.661 | 1 | 0.416 | 2.750 | 1 | 0.097 | 3.439 | 1 | 0.064 | 1.596 | 1 | 0.206 |
| N of valid cases | 150 | 150 | 150 | 150 | ||||||||
Note: df – Degrees of freedom; Superscript ‘a’ indicate 2 cells (20.0%) have expected count less than 5. The minimum expected count was 2.86; ‘b’ indicates 1 cell (10.0%) have expected count less than 5. The minimum expected count was 4.47; ‘c’ indicates 2 cells (20.0%) have expected count less than 5. The minimum expected count was 2.20; ‘d’ indicates 3 cells (30.0%) have expected count less than 5. The minimum expected count was 1.47. There was no significant difference (p>0.05) among age groups regarding respiratory pathogens.
Table (8):
Occurrence of respiratory pathogens among different breeds in eastern plain zone of Uttar Pradesh
| Breeds | Number of samples tested | Pasteurella spp. | Escherichia coli | Klebsiella spp. | Staphylococcus spp. | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number of positive | Percent positivity | Number of positive | Percent positivity | Number of positive | Percent positivity | Number of positive | Percent positivity | ||
| Non-descript | 76 | 21 | 27.63 | 30 | 39.47 | 17 | 22.36 | 8 | 10.52 |
| Jamunapari | 43 | 11 | 25.58 | 19 | 44.18 | 8 | 18.60 | 5 | 11.62 |
| Barbari | 31 | 7 | 22.58 | 12 | 38.70 | 5 | 16.12 | 7 | 22.58 |
| Total | 150 | 39 | 26.00 | 61 | 40.67 | 30 | 20.00 | 20 | 13.33 |
Table (9):
Breed-wise comparison of respiratory pathogens by chi-square test
| Parameters | Pasteurella spp. | Escherichia coli | Klebsiella spp. | Staphylococcus spp. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | df | Asymptotic Significance (2-sided) | Value | df | Asymptotic Significance (2-sided) | Value | df | Asymptotic Significance (2-sided) | Value | df | Asymptotic Significance (2-sided) | |
| Pearson chi-square | 0.297a | 2 | 0.862 | 0.315b | 2 | 0.854 | 0.609c | 2 | 0.737 | 2.920d | 2 | 0.232 |
| Likelihood ratio | 0.302 | 2 | 0.860 | 0.313 | 2 | 0.855 | 0.618 | 2 | 0.734 | 2.625 | 2 | 0.269 |
| Linear-by-linear association | 0.292 | 1 | 0.589 | 0.004 | 1 | 0.950 | 0.598 | 1 | 0.439 | 2.298 | 1 | 0.130 |
| N of valid cases | 150 | 150 | 150 | 150 | ||||||||
Note: df – Degrees of freedom; Superscript ‘a’ indicate 0 cells (0.0%) have expected count less than 5. The minimum expected count was 8.06; ‘b’ indicates 0 cells (0.0%) have expected count less than 5. The minimum expected count was 12.61; ‘c’ indicates 0 cells (0.0%) have expected count less than 5. The minimum expected count was 6.20; ‘d’ indicates 1 cell (16.7%) have expected count less than 5. The minimum expected count was 4.13. There was no significant difference (p>0.05) among age breeds regarding respiratory pathogens.
Table (10):
Occurrence of respiratory pathogens among different seasons of eastern plain zone of Uttar Pradesh
| Season | Number of samples tested | Pasteurella spp. | Escherichia coli | Klebsiella spp. | Staphylococcus spp. | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number of positive | Percent positivity | Number of positive | Percent positivity | Number of positive | Percent positivity | Number of positive | Percent positivity | ||
| Winter | 67 | 18 | 26.86 | 26 | 38.80 | 14 | 20.89 | 9 | 13.43 |
| Summer | 28 | 7 | 25.00 | 12 | 42.85 | 5 | 17.85 | 4 | 14.28 |
| Monsoon | 36 | 11 | 30.55 | 18 | 50.00 | 3 | 8.33 | 4 | 11.11 |
| Post-monsoon | 19 | 3 | 15.78 | 5 | 26.31 | 8 | 42.10 | 3 | 15.78 |
| Total | 150 | 39 | 26.00 | 61 | 40.67 | 30 | 20.00 | 20 | 13.33 |
Table (11):
Season-wise comparison of respiratory pathogens by chi-square test
| Parameters | Pasteurella spp. | Escherichia coli | Klebsiella spp. | Staphylococcus spp. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | df | Asymptotic significance (2-sided) | Value | df | Asymptotic significance (2-sided) | Value | df | Asymptotic significance (2-sided) | Value | df | Asymptotic significance (2-sided) | |
| Pearson chi-square | 1.459a | 3 | 0.692 | 3.073b | 3 | 0.380 | 8.979c | 3 | 0.030 | 0.276d | 3 | 0.965 |
| Likelihood ratio | 1.559 | 3 | 0.669 | 3.142 | 3 | 0.370 | 8.644 | 3 | 0.034 | 0.278 | 3 | 0.964 |
| Linear-by-linear association | 0.230 | 1 | 0.632 | 0.017 | 1 | 0.898 | 0.450 | 1 | 0.502 | 0.000 | 1 | 0.988 |
| N of valid cases | 150 | 150 | 150 | 150 | ||||||||
Note: df – Degrees of freedom; Superscript ‘a’ indicate 1 cell (12.5%) have expected count less than 5. The minimum expected count was 4.94; ‘b’ indicates 0 cells (0.0%) have expected count less than 5. The minimum expected count was 7.73; ‘c’ indicates 1 cell (12.5%) have expected count less than 5. The minimum expected count was 3.80; ‘d’ indicates 3 cells (37.5%) have expected count less than 5. The minimum expected count was 2.53. Except Klebsiella spp. (p<0.05), there was no significant difference among season regarding respiratory pathogens.
Gross pathology of P. multocida infection
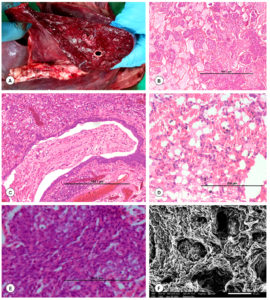
In most cases, pneumonic lesions with consolidation and firm consistency, dark red areas in various lobes especially cranio-ventral lobes were found in the lungs on both sides during necropsy. Focal haemorrhages, congestion, emphysematous patches, fibrinous pleuritis and foamy exudate on cut sections were noticed (Figure 9A). Lungs were grossly heavier than normal with focal or multifocal emphysematous areas and inflated. Hepatized lungs showed diffuse and thickening of the pleura. Further, in majority of the cases, there was noticeable congestion, haemorrhage, and scattered necrotizing lesions in other visceral organs.
Histopathology of P. multocida infection
Microscopically, lung parenchyma of animals showed variable degrees of bronchopneumonia characterized by severe necrosis with nuclear debris and infiltration of neutrophils. Bronchiolar and alveolar lumens were filled with eosinophilic fibrinous and inflammatory cellular exudates, thickened inter-alveolar septa, and engorgement of blood vessels (Figure 9B and C). Bronchiolar epithelium showed degenerative changes. Lungs had severe hemorrhages into the perivascular and alveolar spaces (Figure 9D). The necrotic regions contained spindle-shaped/elongated degenerated leukocytes especially neutrophils and macrophages, which exhibited a streaming pattern of basophilic chromatin known as “oat cells or streaming leukocytes”. Oat cells are also infiltrated in the lumen of alveoli (Figure 9E). In cases of chronic pneumonia, areas of calcification were noticed.
Scanning electron microscopy
The lung tissues from Pasteurella confirmed cases were subjected to scanning electron microscopy (Figure 9F). Walls of the alveoli were lined by a single layer of cells. The affected lungs showed loss of cilia and alveoli were filled with exfoliated cells. Bronchiolar lumen was also filled with exfoliated cells.
Figure 9. Gross and histopathological lesions in P. multocida infection in goats. (A). The cut surface of the lungs showed marked congestion and hemorrhages. (B). Alveolar lumen filled with eosinophilic fibrin and cellular exudates. H&E x100. (C). Bronchiolar lumen filled with eosinophilic fibrin and cellular exudates. H&E x100. (D). Severe hemorrhages in the alveolar lumen and perivascular areas of lungs. H&E x200. (E). Spindle-shaped/elongated necrotic or degenerated leukocytes especially neutrophils and macrophages known as “oat cells”. H&E x400. (F). Lungs showed denudation of alveolar and bronchiolar cells. SEM x50
Gross pathology in E. coli infection
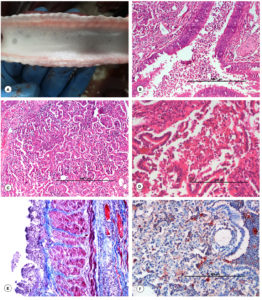
Varied degrees of pulmonary consolidation and pneumonic lesions were evident in the cranio-ventral and cranio-lateral lobes of the lungs on either side along with congestion and hemorrhages. Frothy exudate was observed in the trachea (Figure 10A), bronchi, bronchioles, and cut surfaces of lungs.
Histopathology of E. coli infection
Lungs showed variable degrees of bronchopneumonia. The alveolar lumens were filled with fibrinous exudate, hemorrhages and inflammatory cells especially neutrophils, and severe engorgement of the blood vessels in lungs. Bronchiolar epithelium showed degenerative changes and lumen displayed the infiltration of inflammatory cells and hemorrhages (Figure 10B and C). Alveolar septa showed thickening due to infiltration of inflammatory cells, hemorrhages, and edema. Small intestine showed necrosis, congestion, hemorrhages, and infiltration of inflammatory cells in lamina propria (Figures 10D).
Special staining
In Pasteurella positive cases, the lungs showed the presence of calcium deposits with Von Kossa stain. The mineralized tissues showed an amorphous and black appearance. In E. coli positive cases, the necrotic intestine showed mild to moderate fibrosis with Masson’s trichome stain (Figure 10E).
Immunohistochemistry
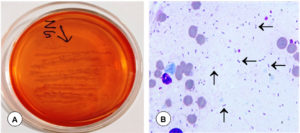
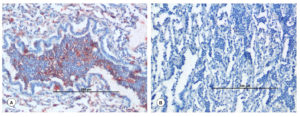
In various tissues, including the lungs and intestine, Pasteurella and E. coli antigens were localized as brown positive immunoreaction. Varied degrees of IHC reaction were observed, which were graded either as mild, moderate, or high, depending on the antigen distribution. In the cases of E. coli infection, most frequently the immunoreaction was found in the cytoplasm of cellular exudates especially in neutrophils and alveolar macrophages, bronchiolar epithelial cells, alveolar interstitial tissues of lungs (Figure 10F), and pulmonary blood vessels. Moderate positive reaction was also seen in the intestinal mucosa. Strong positive immunoreaction was observed in the Pasteurella-infected lungs in the cellular exudates especially in neutrophils and alveolar macrophages and bronchiolar epithelial cells (Figure 11A). The tissue sections from uninfected animals were treated similar to the infected slides and did not show any positive signals (Figure 11B). Antigen distribution typically had a substantial correlation with incidence of pathological lesions. It was interesting to note that there was a correlation between the results of PCR, bacterial isolation, and IHC.
Figure 10. Gross and histopathological lesions in Escherichia coli infection in goats. (A). Trachea showed frothy exudate in their lumen. (B). Bronchiole showed infiltration of inflammatory cells and RBCs, along with degenerative changes in the bronchiolar epithelium. H&E x100. (C). Alveolar lumen contained infiltration of inflammatory cells. H&E x100. (D). Small intestine showed necrosis and infiltration of inflammatory cells in lamina propria. H&E x200. (E). E. coli positive necrotic small intestine showed fibrosis. Masson Trichrome x200. (F). Presence of Escherichia coli antigen in the cytoplasm of alveolar macrophages and neutrophils. IP-DAB-MH, x200
Figure 11. Immunohistochemical and histopathological lesions in P. multocida infection in goats. (A). Presence of Pasteurella antigen in the cytoplasm of alveolar macrophages and neutrophils. IP-DAB-MH, X200. (B). Negative antibody control: Lungs showed no positive immunolabelling for Pasteurella antigen. IP-DAB-MH, X200
Respiratory infections are significantly responsible for the mortality of small ruminants, especially goats.2,32 Respiratory infections must be promptly prevented and controlled to minimize the financial losses to the goat farmers and enhance animal welfare.2,33 The aim of the present study was to report the respiratory infection-causing bacterial pathogens in goats as well as to undertake a molecular and pathological analysis of Pasteurella spp. and E. coli infections. Yun et al.7 reported the high mortality among goats due to respiratory infections and mainly caused by Pasteurella and extraintestinal pathogenic E. coli. But, there are no detailed information regarding the respiratory infection-causing bacterial pathogens in goats of eastern plain zone of Uttar Pradesh, India, available.
The results of bacterial isolation were consistent with the findings reported by Edward and Ewing.10-15 Initial speculation suggested that P. multocida was present in the isolates because that failed to grow on MacConkey agar. However, Mannheimia haemolytica was capable of growing on MacConkey agar, and it was haemolytic, similar results were reported by Quinn et al.20 In the present study, Pasteurella spp. produced non-haemolytic medium-sized, grey, and mucoid colonies with a sweetish odour on blood agar. These findings were in accordance with the reports of Tigga et al.34 and Desam et al.35
In the present study, results of the biochemical test were consistent with the findings of Rawat et al.,36 who reported the positivity for catalase, oxidase, and sucrose tests for both M. haemolytica and P. multocida. P. multocida did not produce hemolysis; whereas, M. haemolytica produces. Voges-Proskauer test was positive for P. multocida; whereas, negative for M. haemolytica. Rawat et al.36 reported that indole test was negative for P. multocida, but positive for M. haemolytica, which was similar to the results of present study. Similar biochemical test findings for E. coli isolated from the respiratory tract of goats were reported by Mahmoud et al.37
The 16S RNA gene sequences of P. multocida isolates showed close similarity (99.69%) with P. multocida strain CCUG 17976, which was identical to the report of Li et al.38 Similar to the report of Davies,39 Pasteurella isolate in the present study showed similarity with P. multocida subsp. gallicida strain CCUG 17978. In the present study, Pasteurella isolate showed similarity with P. multocida subsp. septica strain, which was similar to the earlier reports of Davies39 and Chen et al.40
The percentage prevalence of respiratory infections in different districts of Uttar Pradesh was remarkably comparable to those of Ahmed et al.,41 who reported 24.30% of Pasteurella prevalence. Assefa and Kelkay42 reported 28.4 to 31.4% prevalence of pasteurellosis in goats. Momin et al.43 reported 20% prevalence of P. multocida from black Bengal goats; while, Rashid et al.44 recorded 15% prevalence of P. multocida from the lungs of goats, which were contrast to the present findings. This could be due to variations in the sampling methods, goats with pneumonic diseases, better healthcare and laboratory facilities, ecology of the research locations, and predisposing variable.45 Variation in the results of the current study, compared to the findings of others could be due to most of the researches included both clinically infected and dead animals in their study, but in the present study, only dead goats were included and animals might have been treated with antibiotics for respiratory infections during the clinical course.
In the present study, 20% of samples were positive for Klebsiella spp., and 13.33% of samples were positive for Staphylococcus spp. in the respiratory tract of goats. Klebsiella spp. produces various virulence factors such as lipopolysaccharide, antiphagocytic capsules (K antigen) which aid the bacterium for uptake of iron from the host, and pili for adhesion to host cells. Ibraheim et al.10 reported 35% of positivity to Klebsiella spp. including 30% positivity in trachea, 33.33% in nasal swabs, and 40% in lungs from pneumonic goats in Iraq. Sen et al.12 reported Staphylococcus spp. was the major organism followed by Escherichia coli and Pasteurella spp. in the pneumonia in black Bengal goats.
P. multocida induced pathological findings of the current study were consistent with the findings of Emikpe and Akaptvie46 and Amin.47 Pneumonic lesions with consolidation, firm consistency and dark red areas in various lobes of lungs were the most frequent and conspicuous gross lesions observed in most of the cases. The microscopic lesions observed in the current study were consistent with the earlier report by Hashemnia et al.,48 who reported the presence of fibrin exudate and accumulation of neutrophils in the lumen of the bronchi, bronchioles, and alveoli and fibrinous pneumonia. A rim of elongated cells known as “oat cells” or “swirling macrophages” that formed whorl-like structures in and around the alveoli, which is the characteristic feature of pasteurellosis. These findings were similar to the reports by Singh et al.,4 Azizi et al.49 and Kumar et al.50 Gross and histopathological lesions found in the E. coli-infected lungs were similar to the reports of Singh et al.51 and Sonawane et al.52
In the E. coli-infected intestine, mild to moderate fibrosis was seen and it was evident by Masson’s trichrome staining. In cases of suppurative and chronic caprine bronchopneumonia, regions of calcification were documented earlier by Singh et al.4 In the present study, P. multocida was isolated from the lungs of dead goats, which was in accordance with the findings of Amin.47 Pasteurella spp. antigen can be detected in the formalin-fixed tissue sections by immunoperoxidase staining. Interestingly, in the present study, moderate to strong localization of P. multocida antigen was noticed in the lungs, which was in accordance with the findings of Amin.47 However, a modest response to the IHC may occur due to the low bacterial content in the tissues.
Nevertheless, compared to bacterial culture, the PCR and IHC analysis revealed a higher positive rate of bacterial presence. These results might be explained by the fact that in contrast to the culture approach, which requires living organisms, PCR and IHC can identify nucleic acid and antigen from dead organisms, respectively. In the current study, PCR results for detection of P. multocida and E. coli antigens were inconsistent with the IHC findings. This may be explained by the uneven assimilation of bacteria among the several organs under investigation. Furthermore, presence of lesions often exhibited a substantial association with the localization of P. multocida antigens. As a result, distinct diffuse reactions to P. multocida are probably caused by the bacterial solubilized antigen that results from bacterial degradation and lysis.53 Using IHC, PCR, and bacterial isolation, the current study demonstrated the presence of Pasteurella spp., E. coli, Klebsiella spp. and Staphylococcus spp. antigens in spontaneously infected cases of pneumonia in goats.
Loss of cilia and denudation of cells of alveoli and bronchiole in Pasteurella infected lungs were evident by scanning electron microscopy. These ultramicroscopic lesions in the lungs lead to the failure of gaseous exchange causing the inability of oxygen-carrying capacity in the animal. Therefore, infected animals experience suffocation, and finally, death of animals.
Among the respiratory infections in goats, Pasteurella spp., E. coli, Klebsiella spp. and Staphylococcus spp. infections in eastern plain zone of Uttar Pradesh had been studied limited. Extraintestinal pathogenic E. coli, Pasteurella spp., Klebsiella spp. and Staphylococcus spp. were found in this study. These findings imply that Pasteurella spp. and E. coli infections may be significant contributors to respiratory illnesses in goats. Goats may act as reservoirs for extraintestinal pathogenic E. coli, threatening both public health and goat rearing. The samples crucial for diagnosis of respiratory bacterial pathogens were lungs, followed by heart blood, nasal swabs, and tracheal swabs. The PCR followed by IHC and bacterial culture were the laboratory techniques for diagnosis of respiratory bacterial pathogens. Based on the phylogenetic analysis, most of the isolated Pasteurella strains were having similarities with P. multocida. Results of the present study confirmed the circulation of Pasteurella spp., E. coli, Klebsiella spp. and Staphylococcus spp. among the population of goats in field conditions in eastern plain zone of Uttar Pradesh and should be considered for effective vaccination strategies in small ruminants for the control of respiratory infections.
ACKNOWLEDGMENTS
The authors were highly thankful to the Dean, College of Veterinary Science and Animal Husbandry, Acharya Narendra Deva University of Agriculture & Technology, Kumarganj, Ayodhya, Uttar Pradesh and Joint Director, CADRAD, ICAR-Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh, India.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
DN, SS, and VY conceptualized and visualized the study. DPS performed investigation and data curation. MS and KPS assisted in histopathology, immunohistochemistry and PCR. DN, SS, and VY performed formal analysis. NKD, HK and JKC performed statistical analysis. MS and KPS performed data validation. DPS drafted the manuscript. MS, KPS, NKD, HK and JKC reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
The study was financially supported by the National Agricultural Higher Education Project (NAHEP), Acharya Narendra Deva University of Agriculture & Technology, Kumarganj, Ayodhya, Uttar Pradesh, India, vide grant number NAHEP/IG-2013/2019-20, dated 20.12.2023.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Animal Ethical Committee (IAEC) of Acharya Narendra Deva University of Agriculture & Technology (ANDUA & T), Kumarganj, Ayodhya, Uttar Pradesh, India.
- Dhama K, Chakraborty S, Tiwari R, et al. A concept paper on novel technologies boosting production and safeguarding health of humans and animals. Res Opin Anim Vet Sci. 2014;4(7):353-370.
- Saminathan M, Rana R, Ramakrishnan MA, Karthik K, Malik YS, Dharma K. Prevalence, diagnosis, management and control of important diseases of ruminants with special reference to Indian scenario. J Exp Biol Agric Sci. 2016;4(3S):338-367.
Crossref - Rai RB, Dhama K, Singh B, et al. Impact of novel low cost technological interventions on expenditure pattern of landless and sub-marginal farmers. South Asian J Exp Biol. 2013;3(5):261-267.
Crossref - Singh R, Kumar P, Sahoo M, et al. Spontaneously occurring lung lesions in sheep and goats. Indian J Vet Pathol. 2017;41(1):18-24.
Crossref - Lacasta D, Ferrer LM, Ramos JJ, Gonzalez JM, De las Heras M. Influence of climatic factors on the development of Pneumonia in lambs. Small Rumin Res. 2008;80(1-3):28-32.
Crossref - Milton AAP, Agarwal RK, Priya GB, et al. Captive wildlife from India as carriers of Shiga toxin-producing, enteropathogenic and enterotoxigenic Escherichia coli. J Vet Med Sci. 2019;81(2):321-327.
Crossref - Yun J, Mao L, Li J, et al. Molecular characterization and antimicrobial resistance profile of pathogenic Escherichia coli from goats with respiratory disease in eastern China. Microb Pathog. 2022;166:105501.
Crossref - Sahoo M, Baloni S, Thakor JC, et al. Pathology, virulence-associated gene profiling, antimicrobial susceptibility, and pathogenicity of untypeable capsular serotypes of Pasteurella multocida isolated from slaughtered pigs of India. Lett Appl Microbiol. 2023;76(10):1-9.
Crossref - Chakraborty S, Kumar A, Tiwari R, et al. Advances in diagnosis of respiratory diseases of small ruminants. Vet Med Int. 2014;2014:508304.
Crossref - Ibraheim HK, Madhi KS, Jasim AS, Gharban HAJ. Molecular identification of Klebsiella species from pneumonic goats, Iraq. Open Vet J. 2024;14(11):2980-2988.
Crossref - Mahrous SH, El-Balkemy FA, Abo-Zeid NZ, El-Mekkawi MF, El Damaty HM, Elsohaby I. Antibacterial and anti-biofilm activities of cinnamon oil against multidrug-resistant Klebsiella pneumoniae isolated from pneumonic sheep and goats. Pathogens. 2023;12(9):1138.
Crossref - Sen SK, Chowdhury MR, Mahbub-E-Elahi ATM, Siddique AB. Bacteriological and histopathological investigation of pneumonia in black Bengal goat. Dairy and Vet Sci J. 2018;6(4):555695.
Crossref - Clark SB, Hicks MA. Staphylococcal Pneumonia. In: StatPearls [Internet]. StatPearls Publishing, Florida, 2025.
- Park S, Ronholm J. Staphylococcus aureus in agriculture: Lessons in evolution from a multispecies pathogen. Clin Microbiol Rev. 2021;34(2):e00182-20.
Crossref - Fouad EA, Khalaf DD, Farahat E, Hakim AS. Identification of predominant pathogenic bacteria isolated from respiratory manifested small ruminants in western north Egypt with regard to their susceptibility to antibiotics. Int J Health Sci. 2022;6(S2):10818-10828.
Crossref - Krishnegowda DN, Singh BR, Mariappan AK, et al. Molecular epidemiological studies on avian pathogenic Escherichia coli associated with septicemia in chickens in India. Microb Pathog. 2022;162(2022):105313.
Crossref - Priya GB, Nagaleekar VK, Milton AAP, et al. Genome wide host gene expression analysis in mice experimentally infected with Pasteurella multocida. PLoS ONE. 2017;12(7):e0179420.
Crossref - Shrivastava DP, Niyogi D, Saminathan M, et al. Pathological and immunohistochemical investigation of PPR virus in goats from Ghazipur district of Eastern Plain Zone of Uttar Pradesh. Indian J Vet Pathol. 2023;47(4):292-299.
Crossref - Varshney R, Varshney R, Chaturvedi VK, et al. Development of novel iron-regulated Pasteurella multocida B: 2 bacterin and refinement of vaccine quality in terms of minimum variation in particle size and distribution vis-a-vis critical level of iron in media. Microb Pathog. 2020;147:104375.
Crossref - Quinn PJ, Markey BK, Leonard FC, Fitzpatrick ES, Fanning S, Hartigan P. Veterinary microbiology and microbial diseases. 2nd Edition, Wiley-Blackwell Publisher, UK. 2011;1–928.
- Edward PR, Ewing WH. Identification of Enterobacteriaceae. 3 Edn., Burgees Publishing rd Company, Minnesota, USA. 1972.
- Ingale AM, Rai RB, Saminathan M, et al. Isolation, PCR detection, pathotyping and antibiogram profiling of Escherichia coli associated with endometritis in buffaloes. J Anim Plant Sci. 2016;26(5):1247-1254.
- Srinivasan R, Karaoz U, Volegova M, et al. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS One. 2015;10(2):e0117617.
Crossref - Townsend KM, Hanh TX, O’Boyle D, et al. PCR detection and analysis of Pasteurella multocida from tonsils of slaughtered pig in Vietnam. Vet Microbiol. 2000;72:69-78.
Crossref - Iguchi A, Iyoda S, Seto K, et al. Escherichia coli O-Genotyping PCR: A comprehensive and practical platform for molecular O serogrouping. J Clin Microbiol. 2015;53(8):2427-2432.
Crossref - Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press, New York, USA. 2000.
Crossref - Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series. Sci Res. 1999;41:95-98.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870-1874.
Crossref - Luna LG. Manual of histology staining methods of the Armed Forces Institute of Pathology. 3rd Edition, McGraw Hill, New York. 1968.
- Saminathan M, Singh KP, Maity M, et al. Pathological and immunological characterization of bluetongue virus serotype 1 infection in type I interferons blocked immunocompetent adult mice. J Adv Res. 2021;31:137-153.
Crossref - Jangam AK, Thali P. WASP-Web Agri Stat Package 2.0. ICAR Research Complex for Goa, Ela, Old Goa, Goa, India. 2004;402-403.
- Saminathan M, Singh KP, Khorajiya JH, et al. An updated review on Bluetongue virus: Epidemiology, pathobiology, and advances in diagnosis and control with special reference to India. Vet Q. 2020;40(1):258-321.
Crossref - Rai RB, Dhama K, Chakraborty S, et al. A concept of novel technological approaches for livelihood security and poverty alleviation for poor farmers: a review. Res Opin Anim Vet Sci. 2014;4(7):371-381.
- Tigga M, Ghosh RC, Malik P, Choudhary BK, Tigga P, Nagar DK. Isolation, characterization, antibiogram and pathology of Pasteurella multocida isolated from pigs. Vet World. 2014;7(5):363-368.
Crossref - Desem MI, Ekowati H, Agus S, Safka S, Didik TS, Fitrine E. Morphology, biochemical, and molecular characterization of Pasteurella multocida causing hemorrhagic septicemia in Indonesia. Vet Med Int. 2023;2023(1):1-9.
Crossref - Rawat N, Gilhare VR, Kushwaha KK, et al. Isolation and molecular characterization of Mannheimia haemolytica and Pasteurella multocida associated with pneumonia of goats in Chhattisgarh. Vet World. 2019;12(2):331.
Crossref - Mahmoud MA, Osman WA, Goda ASA, El Naggar AL. Prevalence of some respiratory diseases among sheep and goats in Shalateen, Halaieb and Abu-Ramad areas. Beni Suef Vet Medical J. 2005;15(2):196-202.
Crossref - Li Z, Cheng F, Lan S, et al. Investigation of genetic diversity and epidemiological characteristics of Pasteurella multocida isolates from poultry in southwest China by population structure, multi-locus sequence typing and virulence-associated gene profile analysis. J Vet Medical Sci. 2018;80(6):921-929.
Crossref - Davies RL. Genetic diversity among Pasteurella multocida strains of avian, bovine, ovine and porcine origin from England and Wales by comparative sequence analysis of the 16S rRNA gene. Microbiol. 2004;150(12):4199-4210.
Crossref - Chen HI, Hulten K, Clarridge III JE. Taxonomic subgroups of Pasteurella multocida correlate with clinical presentation. J Clinical Microbiol. 2002;40(9): 3438-3441.
Crossref - Ahmed SJ, Hasan A, Islam MR, et al. Incidence and antibiotic susceptibility profile of Pasteurella multocida isolates isolated from goats in Savar area of Bangladesh. Agricultural Science Digest. 2019;39(4):357-360.
Crossref - Assefa GA, Kelkay MZ. Goat pasteurellosis: serological analysis of circulating Pasteurella serotypes in Tanquaaberegelle and Kola Tembien Districts, Northern Ethiopia. BMC Research Notes. 2018;11(1):1-5.
Crossref - Momin MA, Islam MA, Khatun MM, Rahman MM, Islam MA. Characterization of bacteria associated with pneumonia in Black Bengal goats. Bangladesh J Vet Medicine. 2011;9(1):67-71.
Crossref - Rashid MM, Ferdoush MJ, Dipti M, et al. Bacteriological and pathological investigation of goat lungs in Mymensingh and determination of antibiotic sensitivity. Bangladesh J Vet Med. 2013;11(2):159-166.
Crossref - Abera D, Sisay T, Birhanu T. Isolation and identification of Mannhemia and Pasturella species from pneumonic and apparently healthy cattle and their antibiogram susceptibility pattern in Bedelle District, Western Ethiopia. African J Bacteriol Res. 2014;6(5):32-41.
- Emikpe BO, Akaptvie SO. The clinical and pathological features of experimental Mannheimia hemolytica A2 infection in west African dwarf goats. Bull Anim Health Prod Afr. 2010;58:248-255.
Crossref - Amin A. Pathological investigation on natural infection by Pasteurella multocida in Goats. J Adv Vet Res. 2020;10(2):88-95.
- Hashemnia M, Chalechale A, Malmir E. Pulmonary lesions in slaughtered sheep in Western Iran: gross and histopathological findings. Vet Ital. 2019;55(1):47-56.
- Azizi S, Korani FS, Oryan A. Pneumonia in slaughtered sheep in south-western Iran: pathological characteristics and aerobic bacterial aetiology. Vet Ital. 2013;49(1):109-118.
- Kumar MA, Kumar R, Varshney KC, et al. Pathomorphological studies of lung lesions in sheep. Indian J Vet Path. 2014;38(2):75-81.
Crossref - Singh F, Sonawane GG, Kumar J, Dixit SK, Meena RK, Tripathi BN. Antimicrobial resistance and phenotypic and molecular detection of extended-spectrum b-lactamases among extraintestinal Escherichia coli isolated from pneumonic and septicemic sheep and goats in Rajasthan, India. Turk J Vet Anim Sci. 2019;43(6):754-760.
Crossref - Sonawane GG, Singh F, Tripathi BN, et al. Investigation of an outbreak in lambs associated with Escherichia coli O95 septicemia. Vet Pract. (2012);13(1):72-72.
- Haritani M, Ishino S, Oka M, et al. Immunoperoxidase evaluation of pneumonic lesions in calves naturally infected with Pasteurella haemolytica. Nippon Juigaku Zasshi. 1989;51(6):1137-1141.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.