ISSN: 0973-7510
E-ISSN: 2581-690X
Marine coastal areas of the Red sea in Egypt have great biodiversity of sponges which harbors a huge number of endosymbiotic bacteria which are the source of many potential bioactive metabolites and consider as an excellent source. The study focuses on the isolation of Pseudomonas sp associated with marine sponge Hyrtios aff. Erectus from the Red Sea in pure culture, phylogenetically identified and screening of their metabolic pathways polyketide synthases and Nonribosomal peptides (PSK and NRPs). Characterization of the bacterial produced bioactive metabolite was done through antioxidant and cytotoxicity assays using DPPH and cell line. The 16S rRNA gene sequencing of bacterial isolate revealed that the bacterial strain was closely related to Pseudomonas sp., and the active metabolic pathways genes in the Pseudomonas sp was PSK II. The antioxidants and cytotoxicity bioactivity of Pseudomonas sp as is most probably related to the presence of associated bioactive metabolic pathways genes related to polyketides family type II. Furthermore, the antioxidant activity of Pesudomance sp extract, performed by quantification of total phenolic content (TPC) and 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging activity antioxidant activities, were evaluated. The highest antioxidant activity was exhibited at different concentrations (50 mg-6.25 mg) which showed a potent antioxidant capacity by 100% at 50mg, 25 mg and 12.25mg. From the cytotoxicity, antioxidant and bioactive metabolic screening pathways this bacterial strain was revealed to be an excellent source of natural antioxidant and cytotoxicity compounds against free radical and cancerous cell line.
Pseudomonas sp., Metabolic Pathways NRPs and PKS, Hyrtios aff. Erectus Sponge extract.
Marine microbes are generally exposed to extremes conditions such pressure, temperature, salinity and nutrient availability (Kennedy et al., 2008, De Carvalho et al., 2010, Dalmaso et al., 2015). These harsh conditions possess highly diverse bacterial communities, raising the potentially unique biochemistry of marine microbial environmental habitats (Manivasagana, et al., 2013 El Komy et al., 2015a,b, Abdel Monein et al., 2017).Marine invertebrates are known to be natural unique accumulators of specific microbial communities due to their filter-feeding habits. Additionally, they are an unusually rich source of bioactive natural products with medicinal and pharmaceutical potentials (Taylor et al., 2007, Hegazy et al., 2015 a, b). Furthermore, marine sponges harbor rich communities of symbiotic bacteria that have been reported as the true producers of sponge-derived compounds. Although these bacteria in most cases cannot be cultivated, culture-independent methods, such as isolating and analyzing biosynthetic gene clusters using metagenomic strategies, have recently provided the first insights into their chemical potential (Hochmuth and Piel, 2009).
A tremendous number of biologically active compounds were isolated from marine sponges and their associated microbes (Debbab et al., 2010, El Komy et al., 2015a,b, Abdel Monein et al., 2017). Blunt et al., (2013) reported that about 300 novel compounds were reported in 2011 from the phylum Porifera. Most of sponge-derived products are important with a unique chemically diverse entities and they possess different biological activity such ,as anti-inflammatory, antitumour, immuno-or neurosuppressive, antiviral, antimalarial, antibiotic and antifouling (Imhoff et al., 2011). Which are, in fact, produced by their associated microbial communities (Wang, 2006; Khan et al., 2014). Marine bacteria, were under focuses by researchers as they are attractive because they can potentially produce compounds with unique biological properties (Maithili et al., 2014). Many bacterial isolates were obtained such as marine Streptomyces, Pseudomonas, Pseudoalteromonas, Bacillus, Vibrio, and Cytophaga were isolated from seawater, sediments, algae, and marine invertebrates are known to produce bioactive agents (Soliev et al., 2011). They are able to produce indole derivatives, alkaloids, polyenes, macrolides, peptides, and terpenoids. The marine world, due to its phenomenal biodiversity, is a rich natural resource of many biologically active compounds such as polyunsaturated fatty acids (PUFAs), sterols, proteins, polysaccharides, antioxidants and pigments (Lordan et al., 2011, El-Komey et al., 2015 a, b).
Pseudomonas, on the other hand, are well-known and widespread microorganisms, which have been isolated from a variety of natural sources, soil, plants, mineral waters and clinical specimens. They are characterized by a high level of metabolic diversity (Moore et al., 1996), but the vast majority of species formerly assigned to Pseudomonas and recovered from marine sources are now classified within other genera (Kersters et al., 1996; Brown et al., 2001; Yoon et al., 2003). As reported in many published research papers (Baumann et al., 1983; Rossello´-Mora et al., 1993) few species originating from marine environments are included within Pseudomonas such as Pseudomonas perfectomarina, Pseudomonas alcaligenes, Pseudomonas pseudoalcaligenes and Pseudomonas alcaliphila (Yumoto et al., 2001) from sea waters, and some strains of Pseudomonas bacteria (Bennasar et al., 1996) have been recovered from marine sediments.
Polyketide synthases (PKS) and non-ribosomal peptide synthetases (NRPS) are multifunctional enzymes that are involved in the synthesis of a broad range of structurally diverse natural compounds, many of which are of medical importance (Hutchinson, 2003). Bacteria associated with marine sponge have been shown to produce a huge number of biologically active natural agents, including polyketides (Proksch et al., 2002). Also, many of bioactive compounds that have entered clinical and pre-clinical development are believed to be ultimately microbial in origin isolated from sponge (Kennedy et al., 2008).
Antioxidants are natural substances that may prevent or delay some types of cell damage. These molecules protect the cells from the damage caused by unstable molecules known as free radicals. Marine bacteria have not been examined for antioxidant compounds as extensively as terrestrial bacteria. It was reviewed that the bioactive metabolites extracted from seaweed as well as sponges and their associated bacteria had good antioxidant activity which needs extensive attention in terms of drug discovery (Balakrishnan et al., 2014). Marine sponges, seaweeds and their associated bacteria produce bioactive compounds which have been found to be important for health promotion and disease prevention. These beneficial effects can be attributed to the complex mixture of phytochemicals, which possess antioxidant, antimicrobial, anticancer and antiviral activity. The compounds responsible for these activities comprise phenolic compounds, sulphated polysaccharides and organic acids (Wijffels, 2008). The antioxidants generally produced by marine environment include polyphenolic compounds such as flavonoids, cinnamic acid, benzoic acid, gallic acid, phlorotannins and quercy (Al-Saif et al., 2014).
In the present study, bioactive strain (Pesudomance sp.) was isolated from a sponge specimen and identified by using a phylogenetic tree taxonomic approach. The bacterial strain was investigated for biosynthetic pathways of PSK and NRPs using PCR screening and sequencing. Moreover, the bioactivity of the bacterial strain was screened for their cytotoxicity and total antioxidant capacity using ethyl acetate extracts of bacterial strain against the hepatic cell line.
Sponge Sampling and Identification
Sponge samples were collected from the Red Sea at Hueghada City (Figure 1). The Red sea represents a lot of different environmental ecosystem with more than 10 thousands sea organisms. However, it started to suffer from several major threats (Shriadah et al., 2004; Okbah et al., 2005, Fahmy et al., 2005 & 2016, Abdel-Halim et al., 2007 & 2016; Shreadah et al., 2008a,b; Abo-El khair et al., 2007& 2008 & 2016; Gurguess et al., 2008, Said et al., 2010, Masoud et al., 2010&2012, Shreadah et al., 2008a, b, c, d, 2011, 2015). Samples were collected by scuba diving, transferred directly to the laboratory in sterile polyethylene bags under reduced temperature (zero0C) for identification and microbial isolation. Voucher sample was preserved in 90% ethanol for taxonomic identification and deposited in Naturalis Biodiversity Center, Leiden using general external and internal morphological characters analysis, i.e., shape, type, size and arrangement of skeletal Structures (spicules) following the System Porifera classification system. And further examination of additional samples, enable identification to species level this species was previously reported and identified as Hyrtios aff. Erectus (Figure 2) by Dr. Nicole Voogd, de with voucher number RMNH POR.8633 in the natural biodiversity center Leiden, Holland.
 Fig. 1: The Red Sea map showing the sampling area
Fig. 1: The Red Sea map showing the sampling area Fig. 2: Sponge sample Hyrtios aff. Erectus RMNH POR. 8633 sample
Fig. 2: Sponge sample Hyrtios aff. Erectus RMNH POR. 8633 sampleIsolation and maintaining culture media of bacteria from sponge
The isolation of bacteria from sponge has been done according to method of Santavy et al. (1990). Four different media were applied, namely marine agar media, nutrient agar, R2A and ISP2 agar, all media were supplemented with 0.2 ìm pore size filtered cycloheximide (100 µg/mL), nystatin (25 ìg/mL) and nalidixic acid (25 ìg/mL) to facilitate the isolation of slow-growing strain.
Genomic DNA Extraction
Bacterial cells were collected by centrifugation at 20,000 rpm. Genomic DNA was extracted from 1.75ml of bacterial culture (QIAGEN, USA).
Bacterial identification using 16S ribosomal RNA (r RNA)
The PCR amplification reactions were performed according to Sambrook et al., (1989), using 1 µL of 10 pmol forwarded 16S-rRNA primer (52 -AAATGGAGGAAGGTGGGGAT-32 ); 1 µL of 10 pmol reverse 16S rRNA primer (52 -AGGAGGTGATCCAACCGCA-32 ); The PCR products were cleaned up for DNA sequencing following the method described by Sambrook et al. (1989). Automated DNA sequencing based on enzymatic chain terminator technique, developed by Sanger et al. (1977), was carried out using 3130X DNA sequencer (Genetic Analyzer, Applied Biosystems, Hitachi, Japan).
Phylogenetic analysis for bacterial strain
For bacterial identification, sequences were submitted to BLA-STN (National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov). The 16S rRNA gene sequences from each species and the matched sequences from GenBank were aligned using ClustalW2 similarity analysis of the nucleotides was performed by BLAST searches against sequences available in GenBank. For phylogenetic tree construction, multiple sequences were obtained from GenBank and the alignments were performed using MEGA 5 software version 5.1 (Dereeper et al, 2008, Tamura et al., 2011), also other analysis using geneious 7.0 were done for bacteria.
Polymerase chain reaction (PCR) screening of nonribosomal peptide synthetases (NRPS) and PKS-I, II gene fragments in Pseudomonas bacteria
Screening for biosynthetic genes and analysis of PKS-I, PKS-II and NRPS genes in marine bacterial Pseudomonas (sp) was tested using degenerate PCR primers for the presence of polyketide synthases type I and II (PKS-I and PKS-II) as well as nonribosomal peptide synthetases (NRPS). Ketosynthase (KS) domains of type I polyketide synthase (PKS) gene were PCR amplified from genomic DNA using the primers K1F (52-TSAAGTCSAACATCGGBCA-32) and M6R (52 -CGCAGGTTSCSGTACCAGTA-32 ). Type II PKS sequences were amplified using KSáF (52 -TSGRCTACRTCAACGCSCACGG-32) and KSâR (52 -TACSAGTCSWTCGCCTGGTTC-32). In order to target adenylation domains of NRPS genes, the degenerate PCR primers A3F (52 -GCSTACSYSATSTACACSTCSGG-32) and A7R (52 -SASGTCVCCSGTSCGGTAS-32) were used (Ayuso-Sacido, et al., 2005). Sequences of the corresponding PCR products (KS domains, 1250–1400 bp; KSá and KSâ, 800–900 bp; adenylation domains, 700 bp), (Ayuso-Sacido and Genilloud, 2005) were compared with NRPS and PKS sequences in the NCBI database by using the basic local alignment search tool (BLAST).
PCR products were amplified in each reaction and subjected to electrophoresis using 0.8% agarose gel. Then, each PCR fragment was purified using the QIAquick gel extraction kit (Qiagen). PCR fragments were sequenced by automated DNA sequencing based on enzymatic chain terminator technique developed by Sanger et al. (1977) using 3130X DNA Sequencer (Genetic Analyzer, Applied Biosystems, Hitachi, Japan).
Bioinformatics analysis
The draft genomes were submitted to secondary metabolite gene cluster analysis using PRISM (Skinnider et al.,2016), Nap Dos (Ziemert etal.,2012) and for bioassay analysis we used pub chem Bioassay.
Cultivation, production and isolation of secondary metabolites from bacterial strain for screening purposes
The cultivation and production of secondary metabolites from Pseudomonas sp for in–vitro biochemical assays was done according to the methods of Al-Zereini et al., (2014). Liquid bacterial cultures were grown in 1L erlenmeyer flasks containing 0.5L nutrient broth medium and incubated in shaking incubator at 102 rpm. Culture were homogenized and centrifuged by cooling centrifuge at 8000 rpm for 2 min at 4oC. Resultant cell mass was extracted with EtOAC (3 x 50 ml), the organic fractions were combined, and the solvent removed at reduced pressure at 35oC. Residues were re-dissolved in DMSO for further bioassay studies.
Prescreening bioassays using in-vitro cytotoxicity using cell lines
Different concentrations of Pesudomance marine extracts in ìg/ml were tested for hepatic cell line. Samples were dissolved in DMSO and further diluted with cell culture medium. The final DMSO concentration used was 1% of total volume of the medium in all treatments, including the control group. Cells with no treatment were examined as negative and positive controls, respectively (Kosaniæ et al., 2015).
Determination of total phenolic content in pesudomance marine extracts
Total phenolic contents in the bacterial pesudomance extract was determined by the method of Taga et al. (1984).
Total antioxidant capacity using 2.2.Diphenyle –a-picrylhydrazyl (DPPH) radical scavenging effects of different extracts
DPPH radical scavenging assay of the total extract was performed using modified previously established methodology by Blois (1958) and Amarowicz et al., (2000).. Scavenging ratio of DPPH assay calculated as follows:
% scavenging = [(A control – A sample] / A control x 100
The identification of bacterial strain associated with sponge using 16S rRNA Gene analysis identification
The identification of bacterial strain using 16SrRNA gene analysis using the BLAST search on NCBI resulted in Pseudomonas sp., based on the highest identy value, small gap value (0%), high score, bit and E-value (table 1). As the lower E-value provided the more similar the sequence and the more confidence the BLAST search result which are listed in tables 1 for bacterial strains and shown in phylogenetic tree of Figure (3).
Table (1):
The identification of Pseudomonas sp. using BLAST.
Description |
Max score |
Query cover |
Evalue |
Ident |
Accession |
|---|---|---|---|---|---|
Pseudomonas sp. Nabil 16Sribosomal RNA gene, partial sequence |
1712 |
99% |
0 |
100% |
KR269850.1 |
Pseudomonas pseudoalcaligenes genome assembly Ppseudo_Pac, chromosome : I |
6667 |
99% |
0 |
96% |
LK391695.1 |
Uncultured bacterium clone nbw117a03c1 16S ribosomal RNA gene, partial sequence |
1672 |
99% |
0 |
96% |
KF064855.1 |
Uncultured Pseudomonadaceae bacterium clone S2-8-094 16S ribosomal RNA gene, partial sequence |
1672 |
99% |
0 |
96% |
KF786991.1 |
Uncultured Pseudomonas sp. clone C146500375 16S ribosomal RNA gene, partial sequence |
1672 |
99% |
0 |
96% |
JX531395.1 |
Uncultured Pseudomonas sp. clone C146001084 16S ribosomal RNA gene, partial sequence |
1672 |
99% |
0 |
96% |
JX529416.1 |
Pseudomonas sp. B205-B5 16S ribosomal RNA gene, partial sequence |
1672 |
99% |
0 |
96% |
JX961604.1 |
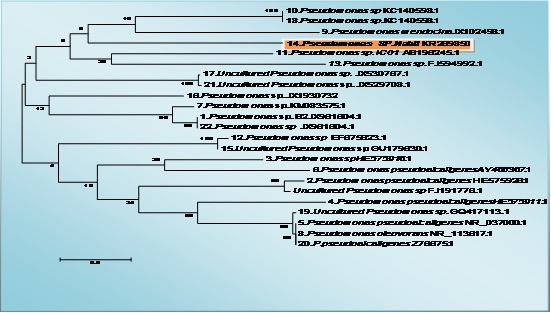
Fig. 3: The tree showing the phylogenetic diversity of pseudomonas sp. with accession number KR269850 Neighbor-Joining (NJ) tree built with MEGA 5.1 based on partial 16S rRNA gene sequences of 23 strains belonging to the pseudomonas sp. and the type strains of closely related genera. The numbers at the nodes indicate bootstrap support (%) based on a NJ analysis of 1.000 replicates; only values ≥75% are given. The scale bar indicates 0.5
Polymerase chain reaction (PCR) screening and sequencing of nonribosomal peptide synthetases (NRPS) and polyketide synthases PKS-II gene fragments in bacteria and Bioinformatics bioactive screening
The Pseudomonas sp. screening to assess their biosynthetic potential pathways were listed and summarized in Table 2 & Fig. 4 and 5. The PCR screening was carried out for the presence of secondary metabolite genes encoding nonribosomal synthetase (NRPS) and polyketide synthases (PKSI&PSKII). The only primer give result was KSF& KSR primer for PKSII while other give –ve result under the same condition for PSK-I and NRPs. The bacterial strain studied was isolated from the Hyrtios aff sponge. Sponges were collected in the coastal area in the Red sea. The gene of 16S rRNA was amplified using the bacterial primers. The obtained sequences were translated in silico into protein sequences using the EMBL program (Hall.,1999) and compared to the published sequence data using the BLAST software of the NCBI site (Altschul et al.,1990).The nucleotide sequences of the 16S rRNA gene and the polyketide synthase KS genes were deposited in the GenBank database (KR269850.1, KU848194.1, AMQ36814). Based on the 16S rRNA gene sequence (~998 bp long), strain KR269850.1, was identified as a Pseudomonas strain; one sequence fragments of ketosynthase (KS) domain genes of the PKS gene cluster were identified in its genome. The amino acid sequences obtained by the in silico translation showed 39% homology. The BLAST analysis of these sequences revealed their similarity to modular KS domains of different bacteria, including known producers of biologically active metabolites and unidentified strains. Their closest homologues, which were identified by a BLAST search, are listed in the Table (2). It was found that KS1_PF and KS2_PF were similar to the KS domains of gene clusters of (uncultured bacterium) Sequence ID: AGG47970.1. the screening of bioactivity using secondary metaboliters software using PRISM, PubMed Chem Bioassay and NaPDoSR resulted in variety of bioactive domain are listed in Tables (3, 4 and 5).
Table (2):
The BLAST result for Pseudomonas bacterial PSK II gene.
Description |
Max score |
Query cover |
E value |
Ident |
Accession |
|---|---|---|---|---|---|
putative type II PKS ketosynthase alpha subunit [uncultured bacterium] |
68.6 |
39% |
5.00E-11 |
50% |
AGG47970.1 |
putative type II PKS ketosynthase alpha subunit [uncultured bacterium] |
68.2 |
38% |
6.00E-11 |
55% |
AGG48159.1 |
putative type II PKS ketosynthase alpha subunit [uncultured bacterium] |
66.6 |
38% |
2.00E-10 |
54% |
AGG47517.1 |
putative type II PKS ketosynthase alpha subunit [uncultured bacterium] |
66.6 |
38% |
2.00E-10 |
54% |
AGG48005.1 |
putative type II PKS ketosynthase alpha subunit [uncultured bacterium] |
66.6 |
38% |
2.00E-10 |
54% |
AGG48003.1 |
putative type II PKS ketosynthase alpha subunit [uncultured bacterium] |
66.6 |
38% |
2.00E-10 |
54% |
AGG47639.1 |
putative type II PKS ketosynthase alpha subunit [uncultured bacterium] |
66.6 |
38% |
2.00E-10 |
54% |
AGG48132.1 |
putative type II PKS ketosynthase alpha subunit [uncultured bacterium] |
66.6 |
38% |
2.00E-10 |
54% |
AGG45514.1 |
Table (3):
The Bioinformatics analysis of bioactive screening using PRISM program for DNA nucleotide sequence of KU848194.1| Pseudomonas sp. Nabil.
PRISM program |
Analyzing domain |
Result |
|---|---|---|
Analyzing adenylation domains |
Finding adenylation domains with substrate valine |
|
Analyzing acyl adenylation enzyme domains |
Finding acyl adenylation enzyme domains with substrate phenyl pyruvate |
|
Analyzing acyltransferase domain |
Finding acyl adenylation domains with Propionyl-CoA |
|
Analyzing substrate biosynthesis domains |
Identifying Aryl formamidase domains |
|
Analyzing other thiotemplated domains |
Identifying starter unit-acyl carrier protein transcylase (C6) domains |
|
Analyzing type II polyketide domain |
Analyzing halogenation and glycosyltransferase domains |
|
Analyzing tailoring domain |
Identifying formyltransferase domains |
|
Analyzing sugar gene |
Identifying UDP-sugar dehydrogenase domains |
|
Analyzing ribosomal domains |
Identifying macrocyclase domains |
|
Detecting clusters |
Clustering biosynthetic orfs… |
|
Cleaving ribosomal precursors |
Predicting RiPP precursor peptide cleavage sites |
Table (4):
The result of bioassay / bioassay protein using pub chem Bioassay (BLASTP 2.5+).
Select for downloading or viewing reports |
Description |
E value |
Ident |
Accession |
|---|---|---|---|---|
Select seq pdb|3I8P|A |
Chain A, Crystal Structure Of E. Coli Fabf (C163a) In Complex With Platensimycin A1 |
3e-13 |
25% |
3I8P_A |
Select seq dbj|BAD72839.1| |
condensing enzyme II [Staphylococcus aureus] |
9e-13 |
25% |
BAD72839.1 |
Select seq ref|WP_000817178.1| |
MULTISPECIES: 3-oxoacyl-ACP synthase I [Proteobacteria] |
1e-07 |
24% |
WP_000817178.1 |
Select seq gb|AAB46389.1| |
fatty acid synthase [Gallus gallus] |
1e-06 |
21% |
AAB46389.1 |
seq Select ref|WP_003411571.1| |
MULTISPECIES: beta-ketoacyl-[acyl-carrier-protein] synthase II [Mycobacterium tuberculosis complex] |
2e-04 |
20% |
WP_003411571.1 |
Select seq ref|NP_004095.4| |
fatty acid synthase [Homo sapiens] |
3e-04 |
18% |
NP_004095.4 |
Select seq ref|WP_003411576.1| |
MULTISPECIES: 3-oxoacyl-ACP synthase II [Mycobacterium tuberculosis complex] |
0.001 |
18% |
WP_003411576.1 |
Select seq emb|CAA44679.1| |
fatty-acid synthase [Rattus norvegicus] |
0.001 |
20% |
CAA44679.1 |
Select seq ref|NP_032014.3| |
fatty acid synthase [Mus musculus] |
0.002 |
19% |
NP_032014.3 |
Table (5):
The result of screening of secondary metabolites using NaPDoSR program.
Query id |
Database match id |
% identity |
align length |
e-value |
pathway product |
domain class |
|---|---|---|---|---|---|---|
gi|1005938447|gb|KU848194.1| |
actinorh_NP_629237_KSa |
46 |
68 |
5e-09 |
actinorhodin |
type II |
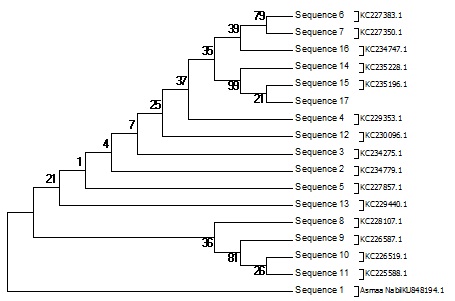
Fig. 4: Phylogenetic diversity Pseudomonas Sp. Neighbor-Joining (NJ) tree based on partial 16S rRNA gene sequences of 17 sequence belonging Biosynthetic gene sequences detected: polyketide synthases PKS-II. The numbers at the nodes indicate bootstrap support (%) based on a NJ analysis of 1.000 replicates; only values ≥75% are given. The scale bar indicates 0.003 substitutions per nucleotide position
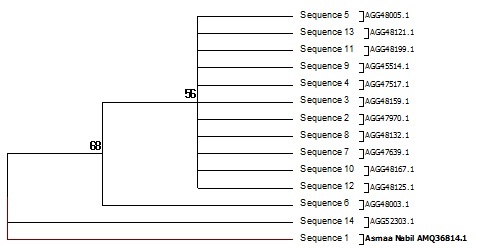
Fig. 5: Phylogenetic diversity Pseudomonas Sp. Tree of the 15 nonredundant protein sequences most identical to type II PKS ketosynthase alpha subunit
Bioactivity of the Pseudomonas Sp. extract
Cytotoxicity of Pseudomonas Sp. extracts against hepatocellular carcinoma cells Inhibitory activity against Hepatocellular carcinoma cells was detected and the IC50 was 31.6 µg.
The total phenolic and total antioxidant capacity
The Pseudomonas extract using ethyl acetate showed higher total phenolic content than water extract as shown in Figure (7). where the levels of total antioxidant capacity in different Psudomance extract concentration have been done using DPPH inhibition. The data in Figure (8) showed the inhibitory effect of Pseudomonas extract at all concentration have been shown potent antioxidant. Where the total phenolic content in ethyl acetate extract showed higher content than water extract as shown in Figure (7).
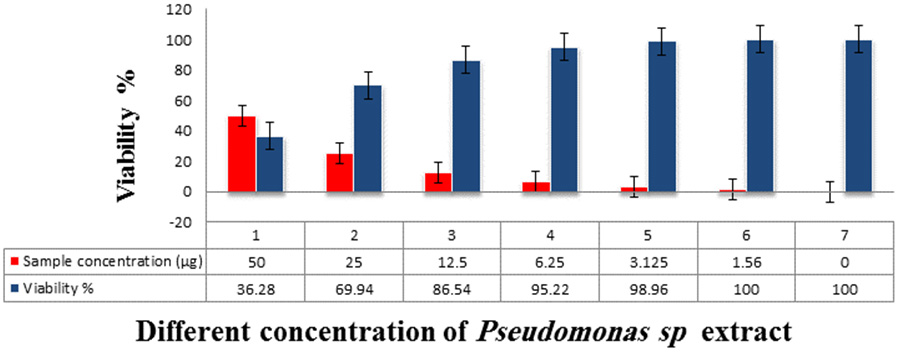
Fig. 6. The Inhibitory activity of Pseudomonas sp. extracts against hepatocellular carcinoma cells
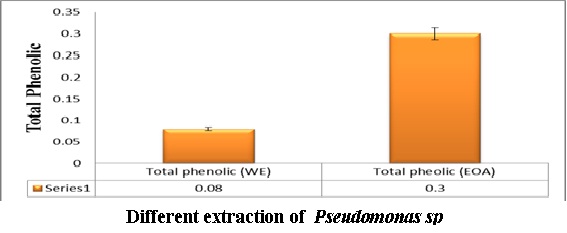
Fig 7: The concentration of total phenolic content in Pseudomonas extract using in water and ethyl acetate extract
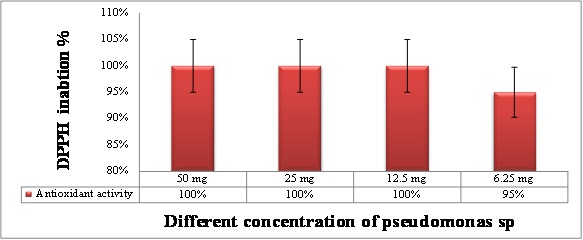
Fig: 8. Total antioxidant capacity in different concentration of Pseudomonas extract using DPPH assay
Marine natural products are valuable sources that could produce potential chemotherapeutic agents; marine derived organisms are widely recognized as important sources of these natural products with interesting biological activities (Graça et al., 2015). Red sea was recognized as a rich source of microbial diversity with unique metabolites that can be of pharmaceutical and medicinal importance. The discovery of novel active metabolites provides an intellectual challenge to examine new products for medicinal purposes (Nadeem et al., 2015).
The sponge-associated microorganisms have revealed high densities, amounting for some sponge species up to 37% of the sponge biomass, which exceeded seawater concentrations by two-third orders of magnitude. More than 30 different phyla have so far been recognized in the domain Bacteria and Archaea as being associated with sponges (Thomas et al.,2010). Environmental stressors such as metal pollution, elevated seawater temperature or other climate changes and/or diseases are speculated as having a significant impact both on the symbiotic microbial community and on the health of sponges (Olson and Kellogg, 2010). Some of isolated bacteria have been specifically associated with marine sponges and are absent from the immediate surrounding seawater, reinforcing the concept of sponge specific microbes (Donia et al., 2003).
In the present study, bacterial strain was identified with potent antioxidant bioactivity as well as high total phenolic content (Figs. 7 and 8) and cytotoxicity against hepatocellular carcinoma cells (Fig. 6). Our result with agreement with many previous studies (Balakrishnan et al.,20104, Horta, et al., 2014, Velho-Pereira et al., 2015, Shindo and Misawa, 2014) which confirmed that the sponge associated bacteria have high bioactive compounds with potent antioxidant and anticancer activity, the bacterial strain which isolated from sponge in our study identified as Psudomance sp in agreement with previous studies (Graça et al.,2015) ,the molecular identification of Pseudomonas sp (GeneBank KR269850) was applied using the partial sequence of 16S rRNA. The bacterial strain was alignment with 23 Pseudomonas sp strains (Fig. 4).The tree topologies were evaluated by bootstrap analyses (Felsenstein, 1985) and phylogenetic trees were inferred using the neighbor-joining method (Saitou and Nei., 1987). The molecular techniques identification depend on the application of PCR that allowed the analysis of small numbers or even single spores and the selection of universal oligonucleotide primers specific to fungi and bacteria. This is facilitated and made the access to nucleotide sequences much easy (Guarro et al., 1999).In the bacterial strain Pseudomonas sp, Gram-negative, aerobic, and non-pigmented, using partial sequence analysis of the 16S rRNA gene showed a high percentage of similarity (96%) to the corresponding sequences of the species Pseudomonas sp. (Fig. 4) in agreement with many papers isolated Pseudomonas sp from sponge (Wang et al., 2014, Ye et al., 2015).
Marine environments and their organisms represent an important source for discovery of new potential active and biologically functional natural products, such as compounds that exhibit anticancer, anti-inflammatory, antiviral and ant-toxic activities (Newman and Cragg, 2010). The purpose of genomic screening work was to screen bacterial isolate Pesudomance sp. which isolated from marine sponge, collected from Red Sea, for potential secondary metabolic production (Ehrenreich et al., 2005). The amplification of genes of PKS-I, PKS-II and NRPS from bacteria were detected using primer (K1f, M6R), (KSá, KSâ) and (A3F, A7R) for PKS-I, PKS-II and NRPS; respectively, only PKSII displayed a positive result. The present results are in agreement with Schirmer et al., 2005 who reported that sponge-associated bacteria are thought to produce many novel bioactive compounds, including polyketides. The majority of the PKS hybridizing clones carried small PKS clusters of one to three modules, although some clones encoded large multimodular PKSs (more than five modules). According to Schirmer et al., (2005) report, the PSK is the most abundant large modular encoded by symbiotic bacteria and that indicate as well as with agreement with our result which confirm this finding as the screening for genomic biosynthetic pathways PKS and NRPs, resulting in a positive result with PSK.
The lack of amplification of NRPs and PSK I genes in bacterial strain pesudomance might indicate their absence or that they were less conserved, hence low homology with the primers. On the other hand, the results of the present study revealing the negative result for NPRS and PKS I genes, although the bacterial strain showed highly bioactivity, suggests that the activities detected possibility suggests that these detected genes may be silent, i.e. nonfunctional (Hutchinson, 2003) and/or the products of these genes may be involved in primary metabolism (Pathom-Aree et al., 2006). Another possible explanation is the fermentation conditions used were not optimal for natural products production.
The present result of the identification of gene clusters potentially encoding secondary metabolites mined using bioinformatics tools, namely PRISM, NapDos, and pub chem Bioassay [Ziemert .,2012, Prlic et al.,2012]. We found a genetic potential of secondary metabolite production in our sequence as many paper reported that the genome Bioactivity potential NRPS and or /PKS is richer and more spread among Gram-negative marine bacteria with genome Bioactivity potential – NRPS/PKS than Alpha- and Gammaproteo bacteria (Machado et al.,2015). Although our collection of secondary metabolites screening using bioinformatics tools is limited in number. Here, we show that using bioinformatics tools a few clusters could be identified, but still the bioactive potential harbored in the genome of these genera appears much lower than that observed in Gammaproteo bacteria (Machado et al.,2015).
The bioactivity of Psudomance sp in the present study as potent antioxidant and cytotoxicity against cancerous cell line can be attributed to the high diversity of marine environmental which lead to great chemidoversty resulted in potent drug candidate (Fenical, 1988). The use of marine natural products as therapeutic and cure tools has been started since 1997 with many discoveries (Fautin, 1988, Cragg and Newman, 2013, Lahlou, 2013). The Psudomance extract was screened against human cancer cell line using the MTT cytotoxicity assay (Fig. 6). The study revealed that marine derived bacteria from the Red sea, especially those belong to genus Psudomance are rich source of structurally novel and biologically potent natural products (Nadeem et al., 2016). In agreement with recent studies which have shown that many microbial extracts from the Red sea displayed significant cytotoxic and apoptotic activity (Blunt et al., 2013, Cragg and Newman, 2013, Chen et al., 2014, Nadeem et al.,2016). Also many paper reported that marine bacteria develop unique metabolic and physiological capabilities, which enable them to survive in extreme habitats and to produce compounds that might not be produced by their terrestrial counterparts which explained potent antioxidant capacity of marine bacteria (Debbab et al.,2010, Kijjoa et al.,2004) another explanation for the highly and unique bioactive compounds from marine bacteria associated with sponge isolated from Red sea was the highly biodiversity of organisms habitat Red sea (Mohamed et al., 2015). As the deep-sea anoxic brines of the Red sea are one of the most extreme environments on earth in comparison to overlying seawater (Elica et al., 2012, Adelmoniem et al., 2013, Mohamed et al., 2015). The anoxic brines are characterized by high salinity (increased from 4% up to 26%), high temperature (up to 70°C), increased concentration of heavy metals, and decrease in O2 levels. Therefore, marine-derived microorganisms have the ability to tolerate and survive in these extreme conditions, since they are uniquely equipped with some self-defence systems. Red Sea microbial diversity (bacteria/fungi) is interesting objects for further studies to discover novel metabolites and new chemotherapeutic agents ( Nadeem et al.,2016).
In conclusion the present study confirmed the highly significance of bacterial associated sponges as reservoir of potent therapeutic and tremendous agents especially as antioxidant . it is consider as the first study in Egypt for our knowledge combine the bioactivity and screening of bioactive metabolic pathway gene as today sponge-associated microbial communities are considered to be responsible for the biosynthesis of many of these agents (anticancer, antioxidant, ant diabetic).
- Abdel-Halim, A. M., Aboel-Khair, E. M., Fahmy, M. A., and Shreadah, M. A. Environmental Assessment on the Aqaba Gulf Coastal waters, Egypt. Egyptian J. Aqu. Res., 2007; 33(1), 1-14.
- Abdel-Halim, A. M., Abdel Nabi, M. A., Abdel Fattah, L. M., Fahmy. M. A., Abo-El-Khair, E. M., khaled, A. M., Abu El-Soud, A. A., and Shreadah, M. A. Environmental studies on the Aqaba Gulf coastal waters during 2011-2013. Journal of Environmental Protection., 2016; 7: 1411-1437.
- Nehad M Abd El-Moneam, Mohamed A Shreadah, Samy A El-Assar, Asmaa Nabil-Adam.(2017).Protective role of antioxidants capacity of Hyrtiosaff. Erectus sponge extract against mixture of persistent organic pollutants (POPs)-induced hepatic toxicity in mice liver: biomarkers and ultrastructural study , HYPERLINK “https://link.springer.com/journal/11356″Environmental Science and Pollution Research, Volume 24, HYPERLINK “https://link.springer.com/journal/11356/24/27/page/1” Issue 27, pp 22061–22072
- Abdel Monein, N. M., Yacout, G.A., Haiam M. Aboul-Ela, H. M., and Mohamed A. Shreadah, M. A. Hepatoprotective Activity of Chitosan Nanocarriers Loaded With the Ethyl Acetate Extract of Astenotrophomonas sp. Bacteria Associated with the Red Sea Sponge AmphimedonOchracea In CCl4 Induced Hepatotoxicity in Rats. Advances in Bioscience and Biotechnology (ABB), 2017; 8(1); 27-50.
- Abo-el-Khair, E. A., Abdel Halim, A. M., Shriadah, M. A. and Fahmy, M. A. Environmental Conditions of the Suez Gulf and the Red Sea Coastal Waters, Egypt. Proceedings of the 8th International Conference on the Mediterranean Coastal Environment. MEDCOAST 2007. E. Ozhan (Editor). 13 – 17 November 2007. Alexandria . Egypt., 2007; P: 517-526.
- Abo-El khair, E. M., Abdel Halim, A. M., Fahmy, M. A., and Shreadah, M. A. Environmental Impact Assessment of Northern Red Sea Regions during 2005 – 2007. Egyptian J. Aqu. Res., 2008c; 34( 2), 20-30.
- Abo-El-Khair, E. M., Abdel Fattah, L. M., Abdel-Halim A. M., Abdel Nabi, M. A., Fahmy, M. A., Ahdy, H. H., Hemeilly, A., Abu El-Soud A., and Shreadah, M. A. Assessment of the hydrochemical characteristics for the coastal waters of the Suez Gulf during 2011-2013. Journal of Environmental Protection, 2016; 7: 1497-1521.
- Azamjon B. Soliev, Kakushi Hosokawa, and Keiichi Enomoto, “Bioactive Pigments from Marine Bacteria: Applications and Physiological Roles,” Evidence-Based Complementary and Alternative Medicine, 2011, Article ID 670349, 17 pages, 2011.
- Balakrishnan.D , Kandasamy.D , Nithyanand.P. A review on Antioxidant activity of marine organismsInternational Journal of ChemTech Research CODEN (USA): IJCRGG ISSN : 0974-4290, 2014; 6(7): pp 3431-3436, Sept-Oct 2014.
- Dalmaso, G. Z. L., Ferreira, D., & Vermelho, A. B. Marine Extremophiles: A Source of Hydrolases for Biotechnological Applications. Marine Drugs, 2015; 13(4), 1925–1965.
- Debbab, A., Aly, A. H., Lin, W. H., & Proksch, P. Bioactive Compounds from Marine Bacteria and Fungi. Microbial Biotechnology, 2010; 3(5); 544–563.
- Debbab A., Aly A.H., Lin W.H., Proksch P. Bioactive compounds from marine bacteria and fungi. Microb. Biotechnol., 2013; 3: 544–563.
- De Carvalho, C. C. C. R., & Fernandes, P. Production of Metabolites as Bacterial Responses to the Marine Environment. Marine Drugs, 2010; 8(3), 705–727.
- Elkomy, R. G., Ibraheem, I. B., Shreadah, M. A. and Mohammed, R. Optimal Conditions for Antimicrobial Activity Production from Two Microalgae Chlorella marina and Nevicula F. delicatula. Journal of Pure and Applied Microbiology, 2015a; 9(4); 2725-2732.
- Elkomy, R. G., Ibraheem, I. B., Shreadah, M. A., Mohammed, R. and Ismael, A. A. Antibacterial and Antifungal Activity of Three Microalgae Isolated from Egyptian Coast of the Mediterranean Sea. Journal of Pure and Applied Microbiology, 2015b; 9(4); 2751-2758.
- Fahmy, M. A., Shriadah, M. A., AbulSoud, A., Abdel Rahman, S. M., and Shindy, M. Hydrography and Chemical Characteristics of the Coastal Water along the Gulf of Suez. Egyptian J. Aquatic Res., 2005; 31: 1-14.
- Fahmy, M. A., Abdel Fattah, L.M., Abdel-Halim, A.M., Abdel Nabi, M. A., Abo-El-Khair,E.M., Ahdy, H. H., Hemeilly, A., Abu El-Soud, A., and Shreadah, M.A. Evaluations of the Coastal Water Quality of the Egyptian Red Sea during 2011-2013. Journal of Environmental Protection, 2016; 7 (12); 1810-1834.
- Gurguess, S. M. Shreadah, M. A., Fahmy, M. A. Aboul El Kheir, E. and Abdel Halim, A. M. Assessment of Water Quality in the Red Sea using in Situ Measurements and Remote Sensing Data. Egyptian J. Aqu. Res., 2009; 35(2); 1-13.
- Hegazy, M. F., Mohamed, T. A., Elshamy, A. I., Hassanien, A. A., Abdel Azimd, N. S., Shreadah, M. A., Abdel gawad, I. I. and El kady, E. M. (2015a). A New Steroid from the Red Sea Soft Coral Lobophytum Lobophytum. Natural Products Research,1-5.
- Hegazy, M. F., Gamal-Eldeen, A. M., Mohamed, T. A., Alhammady, M. A., Hassanien, A. A., Shreadah, M. A., Abdelgawad, I. I. and Elkady, E. M. (2015b). Cytotoxic Constituents from the Red Sea Soft Coral Nephthea Sp., Natural Products Research,1-7.
- Julie B Olson and Christina A. Kellogg.2010. Microbial ecology of corals, sponges, and algae in mesophotic coral environments FEMS Microbiology Ecology Volume 73, Is
- Kennedy, J., Marchesi, J. R., & Dobson, A. D. Marine metagenomics: strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microbial Cell Factories, 2008; 7: 27.
- Kijjoa A., Sawangwong P. Drugs and cosmetics from the sea. Mar. Drugs. 2004; 2: 73–82.
- Maithili S.S , Senthamil. M and , Ramanathan.G.2014. Isolation of Secondary Metabolite From Seawater Bacterial Population and Screening of their Bioactive Potential Against Urinary Tract Pathogens Sourced From HIV Infected Patients, Int.J.Curr.Microbiol.App.Sci, 2014; 3(6): 540-548.
- Mohammed EA (2015). Chemical and biological evaluation of palythoa tuberculosa collected from the red sea. Repository of University of Westerncape.
- Masoud, M. S. Said, T. O., El- Zokm, G. and Shreadah, M. A. Speciation of Fe, Mn and Zn in Surficial Sediments from the Egyptian Red Sea Coasts. Chemical Speciation and Biodiversity, 2010; 22(4); 257-269.
- Masoud, M. S., Said, T. O. El-Zokm, G., and Shreadah, M. A. Assesment of Heavy Metals Contamination in Surface Sediments of the Egyptian Red Sea Coasts. Asturalian Journal of Basic and Applied Sciences, 2012; 6; 44-58.
- Okbah, M. A. Shata, M. A. and Shriadah, M. A. Gochemical forms of trace metals in mangrove sediments-Red Sea (Egypt). Chemistry and Ecology, 2005; 21: 23-36.
- Panchanathan Manivasagana, Jayachandran Venkatesanb, Kannan Sivakumarc, Se-Kwon Kima.2013. Marine actinobacterial metabolites: Current status and future perspectives Microbiological Research, 2013; 168(6); Pages 311–332.
- Said, T. O., Shreadah, M. A., AbdelGhani, S. A. and Abdel Ahmed, A. M. Alkyltin and alkayllead compounds in coastal water of Suez Gulf, Egypt. Egyptian J. Aqu. Res., 2010; 36(1); 33-42.
- Shindo, K., & Misawa, N. New and Rare Carotenoids Isolated from Marine Bacteria and Their Antioxidant Activities. Marine Drugs, 2014; 12(3); 1690–1698.
- Shreadah, M. A., Said, T. O., Abdel Ghani, S. A. and Ahmed, A. M. Distribution of Different Organotin and Organolead Compounds in Sediment of Suez Gulf. Journal of Environmental Protection, 2011; 2(5); 545- 554.
- Shreadah, M. A., Said, T. O., El Zokm, G. and Masoud M. S. Physico-Chemical Characterititics of the Surficial Sediments along the Egyptian Red Sea Coasts. Egyptian J. Aqu.Res., 2008a; 34(4); 16- 34.
- Shreadah, M. A., Said, T. O, Abd El Ghani, S. A., and Ahmed, A. M. Alkyllead andAlkyltin Species in different fishes collected from the Suez Gulf, Egypt. Proceedings of the 2nd International Conference on Aquatic Res, Egyptian J. Aqu. Res., 2008b; 34(4); 64-73.
- Shreadah, M. A. Masoud, M. S., Said, T. O., and El Zokm, G. Application of IR, X-Ray, TGA and DTA to determine the mineral composition of the Sediments and study of reaction kinetics along the Egyptian Red Sea Coasts. Egyptian J. Aqu. Res., 2008d; 34(2), 83- 95.
- Shriadah, M. A., Okbah, M. A., and El-Deek, M. S. Trace metals in the water columns of the Red Sea and the Gulf of Aqaba, Egypt. Water, Air and Soil Pollut., 2004; 153; 115-124.
- Skinnider, M.A., Johnston, C.W., Edgar, R.E., Dejong, C.A., Merwin, N.J., Rees, P.N., and Magarvey, N.A. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proceedings of the National Academy of Sciences, 2016; 113: E6343-E6351. doi: 10.1073/pnas.1609014113.
- Thomas, T. R. A., Kavlekar, D. P., & LokaBharathi, P. A. Marine Drugs from Sponge-Microbe Association—A Review. Marine Drugs, 2010; 8(4); 1417–1468.
- Velho-Pereira, S., Parvatkar, P., & Furtado, I. J. Evaluation of Antioxidant Producing Potential of Halophilic Bacterial Bionts from Marine Invertebrates. Indian Journal of Pharmaceutical Sciences, 2015; 77(2); 183–189.
- Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR The Natural Product Domain Seeker NaPDoS: A Phylogeny Based Bioinformatic Tool to Classify Secondary Metabolite Gene Diversity. PLoS One. 2012; 7(3):e34064 Epub 2012 Mar 29. PubMed PMID: 22479523
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


