ISSN: 0973-7510
E-ISSN: 2581-690X
Brucellosis is an important zoonosis and a significant cause of reproductive losses in animals. Abortion, placentitis, epididymitis and orchitis are the most common clinical manifestations in animals. The present study a total of 168 clinical samples were collected from buffaloes (87) and cattle (81). All clinical samples were processed by cultural isolation on Brucella agar medium (BAM) with selective antibiotic supplements and genus specific PCR using B4/B5 (223bp) and F4/R2 (905bp) primer. Out of 168 clinical samples 15 samples yielded Brucella isolates by cultural isolation and 19 samples positive for Brucella organism by genus specific PCR. All genus specific PCR positive 19 samples also positive by Species specific PCR based on omp31, B. abortus +IS711 (498bp) primer. Amplicon of 498bp by B. abortus +IS711 primers indicates that all nineteen samples of cattle and buffaloes were found to be Brucella abortus.
Brucella abortus, Cultural isolation, Molecular characterization, PCR
Brucellosis is an infectious disease caused by Gram negative, facultative, intracellular bacterial organisms of the genus Brucella that are pathogenic for a wide variety of animals and human beings. The disease has a considerable impact on human and animal health, as well as socioeconomic impacts, especially in which rural income relies largely on livestock breeding and dairy products. It causes significant reproductive losses in sexually mature animals5,27. The disease is manifested by late term abortions, weak calves, still births, infertility and characterized mainly by placentitis, epididymitis and orchitis, with excretion of the organisms in uterine discharges and milk4.
Diagnosis of Brucellosis by cultural isolation, serology and nucleic acid amplification has been explored for the rapid detection and confirmation of Brucella. Cultural isolation and identification of the agent is the gold standard test for Brucella diagnosis, however, this process is risky, time consuming, laborious and not suitable for disease surveillance. Moreover, it is a high risk biological pathogen that requires laboratories with qualified staff and facilities and class 3 personal protective equipment13,22. The Brucellosis diagnosis and surveillance by serological tests, The major drawback of these assays they are not always specific, can cross react with other gram negative bacteria2 and antibodies are not produced at the acute stage of infection17. So, Confirmatory diagnosis by molecular techniques. A number of nucleic acid sequences have been targeted for the development of Brucella genus specific PCR assays, including 16S rRNA24, IS711 genetic element, omp215 and bcsp31.
Collection of Sample
A total of 168 clinical samples like deep vaginal swabs, placental cotyledon, vaginal discharge, aborted fetal materials, milk and blood from cattle and buffaloes.
Isolation
Each sample collected from an animal was separately streaked on Brucella agar medium (BAM) (Hi-media) with selective antibiotic supplements and incubated at 37oC aerobically in an atmosphere of 5 per cent CO2 in CO2 incubator for minimum of 15 days. The plates were observed at every 24 hours interval for the growth. The suspected colonies so obtained were streaked on Blood Agar (BA) and MacConkey Agar (MA).
Identification
The isolates suspected to be of Brucella were subjected to Gram staining and Modified Ziehl-Neelsen (MZN) staining for confirming the purity of cultures and morphological characters. Identification of Brucella organism by agglutination and biochemical test.
Rapid slide agglutination test
One drop (0.03 ml) of known Brucella positive serum (I.V.R.I., Izatnagar) was taken on a glass slide by micropipette. A loopful of culture from suspected single colony was mixed thoroughly with the spreader and then the slide was rotated for four min. The result was read immediately. Definite clumping/agglutination was considered as positive reaction, whereas no clumping/agglutination was considered as negative.
Biochemical characterization of isolates
Oxidase test
Standard oxidase discs (HiMedia Laboratories Ltd., Mumbai) containing 1% NNN’N’ –tetramethyl-p-phenylene diamine dihydrochoride were used to perform the test. The loopful of culture from single colony was just touched on the disc. Development of blue colour within 10 seconds was considered as positive test.
Catalase test
This test was performed by taking 2-3 drops of 3% H2O2 on clean grease-free sterile glass slide and single colony from BAM plate was mixed with the help of a wire loop. Immediate development of gas bubbles was considered as positive test.
Triple Sugar Iron Agar (TSI) Test
In Triple Sugar Iron Agar test a test colony was taken with a sterilized straight inoculation needle and inoculated first by stabbing through the center of the medium to the bottom of the tube and then streaking the surface of the agar slant. Then tube with loose cap was incubated at 350C for 18 to 24 hours and observed for color changes and gas production.
Molecular Detection of Brucella
DNA extraction
Tissues samples were cut into small pieces and triturated along with sterile sea sand in mortar and pestle then homogenates by tissue homogenizer. DNA extraction was carried out from samples using DNeasy Blood and Tissue Kit (Qiagen) following manufacturers protocols.
Detection of Brucella using Genus-Specific B4/B5 primer
A PCR was standardized in a total reaction volume of 25 µl, containing 12.5 µl of 2 x PCR Master mixture, 10 pmol of forward (5’TGG CTC GGT TGC CAA TAT CAA3’) and reverse (5’CGC GCT TGC CTT TCA GGT CTG3’) primers each 1 µl , Template DNA 2 µl and nuclease free water upto 25 µl. The reaction was standardized in a thermal cycler (Eppendorf, Germany). with initial denaturation at 93ºC for 5 min, followed by 35 cycles at 90ºC for 60 s, 64ºC for 30 s and 72ºC for 60 s. Final extension was carried out at 72ºC for 10 min. The amplified product (223 bp) was electrophoresed in 2% agarose gel stained with ethidium bromide (0.5 µg/ml) and image was documented by gel documentation system (Mini BiS BioImaging System).
Detection of Brucella using Genus-Specific F4/R2 primer
A PCR was standardized in a total reaction volume of 25 µl, containing 12.5 µl of 2 x PCR Master mixture, 10 pmol of forward (5’ TCG AGC GCC CGC AAG GGG 3’) and reverse (5’ AAC CAT AGT GTC TCC ACT AA 3’) primers each 1 µl , Template DNA 2 µl and nuclease free water upto 25 µl. The reaction was standardized in a thermal cycler (Eppendorf, Germany). with initial denaturation at 95ºC for 5 min, followed by 30 cycles at 95ºC for 30 s, 54ºC for 90 s and 72ºC for 90 s. Final extension was carried out at 72ºC for 6 min. The amplified product (905 bp) was electrophoresed in 2% agarose gel stained with ethidium bromide (0.5 µg/ml) and image was documented by gel documentation system (Mini BiS BioImaging System).
Detection of brucella using Species-specific B. abortus + IS711 primer
A PCR was standardized in a total reaction volume of 25 µl, containing 12.5 µl of 2 x PCR Master mixture, 10 pmol of forward (5’ GAC GAA CGG AAT TTT TCC AAT CCC 3’) and reverse (5’ TGC CGA TCA CTT AAG GGC CTT CAT 3’) primers each 1 µl, Template DNA 2 µl and nuclease free water upto 25 µl. The reaction was standardized in a thermal cycler (Eppendorf, Germany). with initial denaturation at 95ºC for 5 min, followed by 35 cycles at 95ºC for 90 s, 57ºC for 120 s and 72ºC for 120 s. Final extension was carried out at 72ºC for 5 min. The amplified product (498 bp) was electrophoresed in 2% agarose gel stained with ethidium bromide (0.5 µg/ml) and image was documented by gel documentation system (Mini BiS BioImaging System).
Isolation
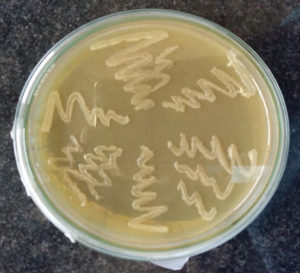
Out of 168 clinical samples, 15 (8.92%) samples produce round, glistening and smooth or mucoid colonies on Brucella agar medium (BAM) (Fig 1, Table 1). These all 15 isolates produce Non-haemolytic colonies on blood agar (BA) but no growth could be obtained on MacConkey agar (MA). In the present finding was in agreement with earlier studies which reported 4% to 8% overall isolation rate6,11. However, in contrast to these findings overall isolation rate between 20 to 39%3,19
Table (1):
Isolation of Brucella On Brucella agar medium
| Animal | Cattle | Buffalo | Total | |
|---|---|---|---|---|
| Vaginal swabs | Tested | 21 | 25 | 46 |
| Positive | 03 (14.28%) | 01 (4.00%) | 04 (8.69%) | |
| Vaginal discharge | Tested | 08 | 06 | 14 |
| Positive | 02 (25.00%) | 01 (16.66%) | 03 (21.42%) | |
| Placenta | Tested | 07 | 06 | 13 |
| Positive | 02 (28.57%) | 01 (16.66%) | 03 (23.07%) | |
| Aborted fetus | Tested | 06 | 11 | 17 |
| Positive | 01 (16.66%) | 04 (36.36%) | 05 (29.41%) | |
| Milk | Tested | 20 | 17 | 37 |
| Positive | 00 | 00 | 00 | |
| Blood | Tested | 19 | 22 | 41 |
| Positive | 00 | 00 | 00 | |
| Total | Tested | 81 | 87 | 168 |
| Positive | 8 (9.87%) | 7 (8.04%) | 15 (8.92%) | |
Identification
Morphological and staining characters of isolates
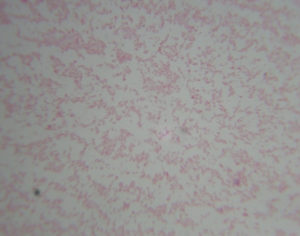
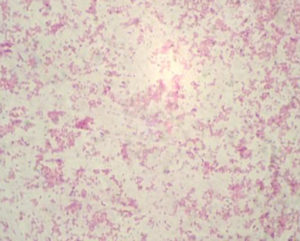
The all 15 isolates were subjected to Gram’s staining and Modified Ziehl-Neelsen’s (MZN) staining. In Gram’s staining pink, gram negative, coccobacillary rods (Fig 2). while in MZN staining they appeared to be red coccobacillary organisms (Fig 3). Similarly, morphology of organism observed by some other authors1,6,11,19.
Rapid Slide Agglutination Test
All the colonies presumed to be of Brucella organism were tested for agglutinatibility with known positive anti Brucella serum. All the isolates revealed clear agglutination, indicative of Brucella abortus.
Biochemical characterization of isolates
All these 15 isolates gaved positive reaction in Catalase (Fig 4) and Oxidase test (Fig 5). On TSI slant, organism showed reaction as Slant (yellow), Butt (black) indicative as Brucella abortus (Fig 6). Pal and Jain (1985)19 and Rhyan et al. (1994)23 reported catalase and oxidase positive for B. abortus.
Molecular Detection of Brucella
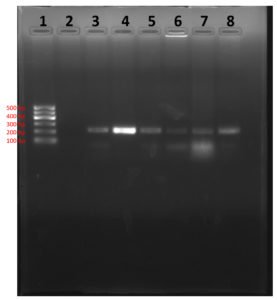
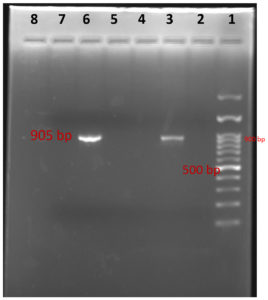
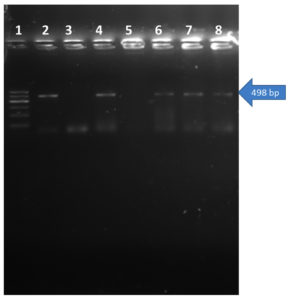
In PCR study targeting 16S rRNA gene, Out of 168 clinical samples nineteen samples were found positive to give specific amplicon of 223bp region of the sequence encoding a 31 kDa immunogenic bcsp31 by Brucella genus specific primer pairs B4/B5 (Fig 7) and 905bp region of the sequence 16S rRNA of B. abortus by Brucella genus specific primer pairs F4/R2 (Fig 8). All genus specific positive nineteen samples yielded an amplicon of 498bp in +IS711 primers indicate species as Brucella abortus (Fig 9). Similarly, Kanani (2007)9 and Jung et al. (1998)11 detection of Brucella by using bcsp31 gene based B4/B5 primer. Navarro et al. (2002)18 and Varasada (2003)26 using same primer pair for diagnosis of human brucellosis. Earlier Navarro et al. (2002)18, Kanani (2007)9 and Patel (2007)21 used same three primer pairs for molecular detection of Brucella abortus. Patel et al., (2015)22 and Karthik et al., (2014)12 used species specific +IS711 primers for detection of Brucella abortus and they yielding 498 bp band when electrophoresed through 2 per cent agarose gel.
1- Ladder (100 bp); 2-Negative Control
3- Positive Control; 4- Sample (positive)
5-Sample (positive); 6- Sample (positive)
7- Sample (positive); 8- Sample (positive)
Fig. 7. Agarose gel showing genus specific PCR amplification products of 223bp by using primer pairs B4/B5 (bcsp31)
1- Ladder (100 bp); 2- Negative control
3- Sample (positive); 4- Sample (negative)
5-Sample (negative); 6- Sample (positive)
7- Sample (negative); 8 -Sample (Negative)
Fig. 8. Agarose Gel electrophoresis of showing genus specific PCR amplification products of 905bp with F4/R2 primers
1- Ladder (100 bp); 2-Positive Control
3- Negative control; 4- Sample (Positive)
5-Sample (Negative); 6,7,8- Samples (Positive)
Fig. 9. Agarose Gel electrophoresis of showing species specific PCR amplification products of 498 bp PCR product with primer IS711
Comparative evaluation of cultural and molecular methods for detection of brucella infection
A total 168 clinical samples are collected for cultural isolation and molecular detection of Brucella organism by PCR. Of these, 19 samples detected positive for Brucella which were further identified as B. abortus. When these samples were processed for the isolation only 15 samples yielded B. abortus.8,10,14,16,25 Detected more number of positives samples by PCR assay than cultural methods.
Present study indicated that B. abortus is widely prevalent in five districts of Gujarat (viz. Banaskantha, Patan, Sabarkantha, Surat and Katchchh) as a cause of Bovine Brucellosis. The isolation results showed the presence of B. abortus in clinical samples which is of public health importance because it is zoonotic disease. There is need to educate about how to prevent and control of brucellosis due to it cause high socioeconomic loss to the farmer.
- Alton, G. G., Jones, L. M., Angus, R. D. Verger, J. M. Techniques for the brucellosis laboratory (1St ed). Institute National Recherche Agronomique (INRA), Paris. 1988; Pp 190.
- Corbel, M. J. and Brinley-Morgan, W. J. Genus Brucella. Meyer and Shaw 1920, 173AL, In Krieg, N.R., Holt, J.G.: Bergey’s Manual of Systematic Bacteriology, Vol.1, Williams & Wilkins, Baltimore-London, 1984; 377-388.
- Das, V. M., Paranjape, V. L., Corbel, M. J. Investigation of brucellosis-associated abortion in dairy buffaloes and cows in Bombay. Indian Journal of Animal Science. 1990; 60(10): 1193-1194.
- England, L., Kelly, R. D., Jones, A., MacMillan, M., Wooldridge. A simulation model of brucellosis spread in British cattle under several testing regimes. Preventive Veterinary Medicine. 2004; 63: 63–73.
- Forbes, L.B. and Tessaro, S.V. Experimental Brucella abortus infection in wolves. Journal of wildlife diseases., 1996; 40(1): 60-65.
- Ghodasara, S. Serological, cultural and molecular characterization of reproductive disorder in various animals and serodetection of brucella antibody. An M.V.Sc. thesis submitted to A. A. U., Anand. 2008
- Halpern, S.D., Ubel, P.A., Caplan, A.L. Solid-organ transplantation in HIV-infected patients. N. Engl. J. Med., 2002; 347(4): 284-7.
- Ilhan, Aksakal, A., Ekin, I. H., Gulhan, T., Solmaz, H., Erdenlig, S. Comparison of culture and PCR for the detection of Brucella melitensis in blood and lymphoid tissues of serologically positive and negative slaughtered sheep. Letters in Applied Microbiology. 2007; 46(3): 301-306.
- Jung, S.C., Jung B.Y., Woo, S.R., Cho, D.H., Kim, J.Y., Kim, W.T., Lee, J.M., Park, Y.H., Baek, B.K. (). Development of a PCR assay for the detection of Brucella spp. in bovine semen. Korean Journal of Veterinary Research. 1998; 38: 345-352.
- Kala, M. Serological, cultural and molecular methods for detection of Brucella infection of bovines in North Gujarat. A thesis submitted to S. D. A. U., S.K. Nagar. 2009
- Kanani, A. N. Serological, cultural and molecular detection of Brucella infection in breeding bulls. Ph. D. thesis submitted to A. A. U., Anand. 2007
- Karthik, K., Rathore, R., Thomas, P., Arun, T.R., Viswas, K.N., Agarwal, R.K., Manjunathachar, H.V., Dhama K. . Loop-mediated isothermal amplification (LAMP) test for specific and rapid detection of Brucella abortus in cattle. Veterinary Quarterly. 2014; 34(4): 174-179.
- Lage, A.P., Poester, F.P., Paixão, T.A., Silva, T.A., Xavier, M.N., Minharro, S., Miranda, K.L., Alves, C.M., Mol, J.P.S., Santos, R.L. Brucelose bovina: uma atualização. Revista Brasileira De Reproduo Animal. 2008; 32(3): 202–212.
- Lavaroni, O., Aguirre, N., Vanzini, V., Lugaresi, C., Torioni de Echaide, S. Assessment of polymerase chain reaction (PCR) to diagnose brucellosis in a Brucella infected herd. Revista Argentina de Microbiología. 2004; 36:101-106.
- Leal-Klevezas, D. S., Martinez, V. I. O., Lopez, M. A., Martinez, S. J. P. Single step PCR for detection of Brucella spp. from blood and milk of infected animals. Journal of Clinical Microbiology. 1995; 3: 3087-3090.
- Leyla, G., Kadri, G., Umran, O. Comparison of polymerase chain reaction and bacteriological culture for the diagnosis of sheep brucellosis using aborted fetus samples. Veterinary Microbiology. 2003; 93(1): 53-61.
- Moussa, I. M., Omnia, M. E., Amin, A. S., Ashgan, M. H. and Selim, S. A. Evaluation of the currently used polymerase chain reaction assays for molecular detection of Brucella species. African Journal of Microbiology Research. 2011; 5: 1511-1520.
- Navarro, E., Escribano, J., Fernandez, J. A. and Solera, J. Comparison of three different PCR methods for detection of Brucella spp. in human blood samples. FEMS Immunology and Medical Microbiology 2002; 34: 147-151.
- Pal, M and Jain, H. S. Investigation into an outbreak of abortion in buffaloes due to Brucella abortus. The Indian Journal of Animal Research. 1985; 6: 37-34.
- Patel, B.C., Chauhan, H.C., Chandel, B.S., Dadawala, A.I., Jain., B.K. Seroprevalence and Molecular characterization of Brucella spp. in buffalo from North Gujarat, India. International Journal of Current Microbiology and Applied Sciences. 2015; 4(4): 174-180.
- Patel, T. J. Serological, cultural and molecular detection of Brucella infection in bovines including quantification in milk by real-time PCR. An M.V.Sc. thesis submitted to A. A. U., Anand. 2007
- Poester, F., Figueiredo, V.C.F., Lobo, J.R., Gonçalves, V.S.P., Lage, A.P., Roxo, E., Mota, P.M.P.C., Müller, E.E., Ferreira Neto, J.S. Estudos de prevalência da brucelose bovina no âmbito do Programa Nacional de Controle e Erradicação de Brucelose e Tuberculose: Introdução. Arquivo Brasileiro de Medicina Veterinária e Zootecnia. 2009; 61(1): 1–5.
- Rhyan, J.C., Quinn, W.J., Stockhouse, L.S., Henderson, J.J., Ewalt, D.R., Payer, J. B., Johnson, M., Meagher, M. Abortion caused by Brucella abortus Biovar 1 in a free-ranging bison (Bison bison) from yellowstone National Park. J. Wildl. Dis. 1994; 30 (3): 445–446.
- Romero, C., Pardo, M., Grillo, M. J., Diaz, R., Blasco, J. M., Lopez-Goñi, I. Evaluation of PCR and indirect enzyme-linked immunosorbent assay on milk samples for diagnosis of brucellosis in dairy cattle. Journal of clinical microbiology. 1995; 33(12): 3198-3200.
- Stella, M., Constantine, A., Efimia, S., Eudoxia, D., Athina, K. Evaluation of different PCR assays for early detection of acute and relapsing brucellosis in humans in comparison with conventional methods. Journal of Clinical Microbiology. 2007; 45(4): 1211-1218.
- Varasada, R. N. Seroprevalence of brucellosis in cattle, buffalo and human being in central Gujarat. A M. V. Sc. thesis, submitted to Gujarat Agricultural University, SardarKrushinagar, India. 2003
- Wadood, F., Ahmad, M., Khan, A., Gul, S. T., Rehman, N. Seroprevalence of brucellosis in horses in and around Faisalabad. Pak. Vet. J., 2009; 29:196-198.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.