ISSN: 0973-7510
E-ISSN: 2581-690X
Keratinolytic microorganisms are highly valuable for decomposition of poultry waste. This study aimed to isolate keratin-decomposing actinobacteria from poultry farm soils and examine their capacity to decompose feathers. Soil samples were placed in a basal medium with feather meal, which is a deposit of carbon and nitrogen. Nine actinobacterial strains were isolated. Actinobacteria were cultured in the media to show clear feather-decomposing potential. Actinobacterial strains were identified using 16S ribosomal RNA (rRNA) sequencing as being related to Streptomyces rochei AM8. Thus, the supernatant of S. rochei AM8 exhibited keratinolytic enzyme activity. Increased biodecomposition of feathers was recorded in a keratinase assay (0.782 U/mL) for separated cultures. The ability of the selected microorganisms to decompose feathers may be an effective biotechnological solution for managing feather waste from poultry.
Feather-degrading Actinobacteria, Characterization, Identification, Keratinase, Feather Waste
There is a significant ecological impact from the increased production and consumption of poultry (including the use of feathers) due to improper waste management strategies.1 Decomposing feathers is naturally slow; moreover, degradation leads to the release of sulfur compounds that are highly toxic to local ecology. The composition of feathers is 90% keratin. Moreover, feathers are extensively deposited by poultry-processing factories and are exploited as waste by-products.2 Unlike other protein macromolecules, keratin is physically insoluble, which means it has incredibly slow decomposition dynamics.2,3 Several chemical and physical techniques are available to transform feathers into digestible proteins that can be used as dietary livestock feed. Nevertheless, processing destroys vital amino acids, reducing the overall quality and digestibility of protein.3 Recently, the alternative manufacturing of microbiota keratinases has become prevalent in the biotechnological industry.
The biological decomposition of keratin is achieved with the help of keratinase, an extracellular enzyme. Keratinase splits the disulfide bonds of keratin to stimulate its decomposition. Several microbes may generate keratinases when a keratin substrate is available. The bacteria responsible for generating keratinase can decompose chicken feathers, hair, nails, and wool. Keratinolytic enzymes are prevalent in the natural environment, as they are released by organic microorganisms. Thus, several bacteria may produce such enzymes, including: Bacillus sp.,3,8 Fervidobacterium islandicum,4 Elizabethkingia meningoseptica KB0425, and Pseudomonas aeruginosa KS16. Actinomycetes such as Streptomyces sp.6-8 Moreover, some fungi may share this capability, such as: Chrysosporium tropicum,9 Trichoderma atroviridae,1 Doratomyces microspores,11 Paecilomyces marquandii,12 Scopulariopsis brevicaulis,13 Alternaria sp., Paecilomyces sp., Penicillium sp., Curvularia , and several Aspergillus sp.14
The variety of keratinolytic fungi found in soils has been thoroughly examined and recorded.15,16 Therefore, keratinases released by keratinolytic microorganisms may be exploited for the decomposition of feather waste. Additionally, digestion products are useful resources for the manufacture of animal feed, fertilizers, and natural gas.17 Relying on keratinolytic organisms is an efficient, cost-preserving, and ecologically sustainable alternative strategy for decomposing feather waste. Similarly, keratinolytic proteases provide a promising opportunity for using low-cost biological technology to eliminate pollutants and manufacture nutritionally augmented protein-saturated feed for cattle.18 Currently, burning and landfilling are conventional methods for removing feather waste that are expensive and toxic to local ecology and contaminate water, soil, and air. Relying on feathers as a fermentation substrate along with keratin-decomposing organisms and enzymatic decomposition may be a better option, thus enhancing the nutritional value of feathers from poultry processing and minimizing the effect on ecology from waste management.19 Such an updated concept may also help to manage the issue of the disposal of poultry-related waste, as keratinaceous waste recycling may be a cost-efficient and ecologically sustainable solution.
The method of submerged fermentation applied to poultry waste, involving organisms that release keratinase, contributes to the transformation of non-soluble keratin present in feathers into solvable proteins or, alternatively, polypeptides.20 The by-products of proteins can be utilized as feed for livestock and leather production.21 Keratinase has commercial value for de-hairing activities in the leather manufacturing industry and is a beneficial substitute for sodium sulfides.22 This study aimed to isolate, define, and describe keratin-decomposing microorganisms obtained from the farming soil of a poultry plant and investigate their feather-decomposing capacities.
Intensified keratinolytic dynamics mostly refer to thermophilic microorganisms that require high-energy sources to achieve the highest growth and degeneration activities.23,24 Actinobacterial isolates can decompose unprocessed feathers. They are also helpful in activating vital processes using keratin substrates. The current research reflects on the extraction and analysis of soil containing feathers, the isolation of actinobacteria from extracted soil samples, and the identification of keratinolytic actinobacterial isolates through primary as well as secondary screening tests.25,26
Obtaining Soil Samples
Actinomycetes were separated and isolated from selected soil samples using a specific approach.27 The samples were obtained from different locations, including compost pits, dumping sites for hair and feathers, and city waste sites in Jeddah, Saudi Arabia. The top layers of the soil were removed (8–10 cm depth) using a clean spade to obtain soil samples with a clean plastic spoon. Soil samples of approximately 50 g were placed in sterile conical containers, which were further labeled with principal information, such as the date, site of collection, and sample number. All samples were stored at 4°C.
Actinomycetes Isolation
The soil samples were processed in broth containing powdered feather meal. The composition (g/L) was as follows: NH4Cl, 0.5; NaCl, 0.5; K2HPO4, 0.3; KH2PO4, 0.4; MgCl2•6H2O, 0.1; and feather powder, 10.0 (pH 7.5). Importantly, the feathers were marked as a source of carbon, nitrogen, and sulfur. The cultures were incubated at 30°C at a selected spinning speed (200 rpm) for 7–10 d. All samples were analyzed in triplicate.
Keratinase Analysis
Keratinase capacities were measured using the approach of Letourneau et al. 28, with some modifications. Keratin azure (Sigma-Aldrich, USA) was used as the substrate. The reaction mixture used in the analysis incorporated 1 mL of keratin azure suspension and 1 mL of untreated enzyme. Reactions were recorded at 50°C in a water bath for half an hour. At the end of incubation, the reactions were stopped using 2 mL of 0.4 M trichloroacetic acid (TCA) to initiate further centrifugation at 3000×g for 5 min with the intention of deleting the substrate. The supernatant was spectrophotometrically used to calculate the release rate of the azo dye at 595 nm. One unit (U) of keratinase was characterized as the quantity of enzyme provoking a 0.01 absorbance elevation between the sample and control specimen at 595 nm in terms of selected conditions. All samples were analyzed in triplicate.
Morphological Description
A morphological analysis based on using the substrate mycelium, aerial mycelium, sporulation, and pigmentation status of actinomycetes29 after proper growth and development on starch casein agar was completed following the steps provided in Bergey’s Manual of Systematic Bacteriology.30 The development parameters of the aerial mycelium, pigmentation dynamics, and growth activity of all 20 isolates were examined. Then, DNA sequencing was performed. The sequences measured were contrasted with those presented in the GenBank network at the National Center for Biotechnology Information (NCBI) using BLAST software.
Amino Acid Assay Using TLC
Amino acid mixtures were isolated on the surface of chromatographic paper (TLC).40 The isolated acids were visualized using a ninhydrin-acetonic solution. A purple color was observed during the reaction between amino acids and ninhydrin.
Effect of pH and Temperature on Keratinase Production
The effects of temperature and pH on enzyme production were examined. Approximately 2 mL of actinomycete isolates (4×104 colony-forming units [CFU]/mL) were inoculated in 250 mL Erlenmeyer flasks containing 50 mL of previously made keratin broth. Afterward, the flasks were incubated at various temperatures and pH levels for a week under specific agitation (200 rpm). At the final stage of the growth process, growth dynamics and keratinase performance of the inoculated isolates were recorded and calculated. All samples were analyzed in triplicate.
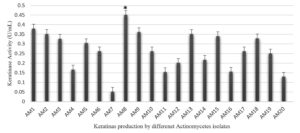
Twenty soil bacterial isolates were examined, with a focus on keratinase-generating microorganisms. The isolates showing moderate to high keratinolytic activity were tested for keratinase enzyme production in basal media (Figure 1). Isolate AM8 showed the highest keratinase activity in the basal media, reaching 0.451 U/mL (Figure 2).
Figure 1. Keratinase production (U/ml) by different Actinomycetes isolates in the basal feather medium
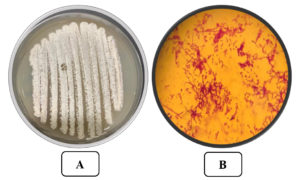
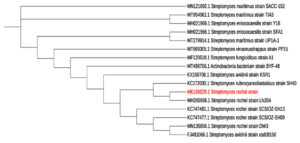
Isolate AM8 was marked and described by defining its morphological, cultural, and biochemical qualities. Isolate AM8 was labeled as a gram-positive, aerobic, non-motile organism with an irregular shape (Figure 3) incapable of developing at 15°C (Table 1). These microorganisms have been found to be moderately tolerant to high temperatures (up to 50°C), with a temperature of 35°C being optimal. Isolate AM8 exhibited stable growth rates at pH levels between 5 and 9 (pH 7 was found to be optimal). Regarding biochemical qualities, these microorganisms have been identified as catalase-positive and oxidase-positive. The morphological parameters of the selected and isolated AM8 actinomycetes are listed in Table 2. The biochemical characteristics of AM8 were successfully defined and are listed in Table 3. The isolate was classified under the genus Streptomyces based on its biochemical parameters. Based on 16S rDNA genotyping and phylogenetic assays, AM8 was identified as S. rochei AM8 (Figure 4).
Table (1):
Morphological characters of the selected actinomycete isolate AM8.
Character |
AM8 |
|---|---|
Source of isolation |
Soil |
Gram stain |
+ve |
Acid fast stain |
-ve |
Shape (Colonies) |
Irregular |
Color |
Pink |
Motility of spore |
Not motie |
DNA G+C content (mol%) |
70-74 |
Spore chain |
Rectifiable |
Spore surface |
Smooth |
Aerial hyphae |
+ |
Substrate mycelium |
+ |
Table (2):
Morphological characters of the selected actinomycete isolate AM8 (ISP).
Media |
Growth |
Color of aerial mycelium |
Color of mycelium substrate |
Presence of soluble pigment |
|---|---|---|---|---|
Starch-nitrate agar |
Heavy |
White |
White |
Pink |
Yeast extract-malt extract agar (ISP-2) |
Heavy |
Pinkish |
Beige |
No pigment |
In-organic salts-starch iron agar (ISP-4) |
Moderate |
Pink |
Beige |
Pink |
Glycerol asparagine agar (ISP-5) |
Moderate |
White |
White |
No pigment |
E- Medium (ISP-9 ) |
Low |
White |
White |
No pigment |
Table (3):
Selected physiological and chemical characters of the selected actinomycetes isolate AM8.
Character |
AM1 |
|---|---|
Growth below 50°C |
+ |
Catalase |
+ |
Oxidase |
+ |
Glycogen |
+ |
L-Histidine |
+ |
L-Proline |
+ |
L-Tryptophan |
+ |
Urease |
+ |
keratinase |
+ |
Cell wall type L-DAP |
+ |
Glycine |
+ |
Arabinose |
– |
Figure 3. A) The Streptomyces rochei AM8 on Starch agar medium B) The Streptomyces rochei AM8 at x 1000 after Gram staining
Figure 4. Partial 16S rDNA gene sequences based on phylogenetic tree of Streptomyces rochei AM8 (acc. no.MK166029.1) and closely associated strains
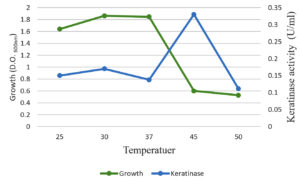
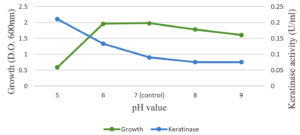
The optimal temperature settings and pH levels for keratinase activity were successfully identified (Figure 5). The optimal temperature for keratinase activity, 30–50°C at pH 5.0, varies depending on the reaction temperatures. In addition, 45°C was defined as the optimal temperature for proper keratinase performance. When the temperature was above 50°C, the dynamics were dramatically reduced (Figure 4). Temperatures above 60°C led to the total deactivation of enzyme productivity. The optimal pH level for keratinase performance was found 5.0, whereas the enzyme productivity decreased dramatically at pH levels above 6.0 (Figure 6).
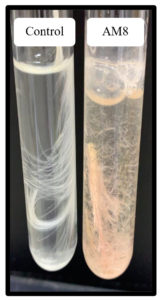
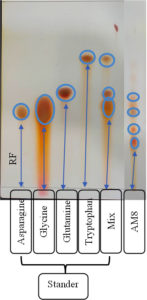
Referring to the enzyme productivity of actinomycetes, S. rochei AM8 was selected for TLC analysis. Keratinolytic enzymes associated with S. rochei AM8 have been exploited to decompose feather molecules and remove large amounts of amino acids. TLC has been utilized to identify free feather-related amino acids produced by enzymatic decomposition reactions. Overall, nine amino acids were identified in the feather samples after degradation. These amino acids, along with critically important amino acids such as arginine, cysteine, asparagine, leucine, tryptophan, tyrosine, glutamic acid, lysine, and alanine, exhibit diverse colors on silica gel with ninhydrin as a mobile stage (Figure 7). In addition, feather-related lysates were evaluated for the release of free amino acids during feather decomposition. The presence of other critically important amino acids, namely leucine and isoleucine, shows that processing with the help of keratinolytic actinomycetes, such as S. rochei AM8, leads to the formation of a feather meal with elevated nutritional value. The present study indicates that feather degradation by S. rochei AM8 is economical and a viable process for better utilization of much-neglected feather waste.
Feather decomposition by 92% with the help of isolated S. rochei AM8, also outside of optimal conditions, may be useful to extensively decompose feathers. In soils, feathers are decomposed by a range of bacteria and fungi. These microorganisms may compete for keratin products or function synergistically. Biological decomposition by organisms incorporating keratinolytic capacities provides an opportunity for alternative strategies to strengthen the nutritional value of keratin waste because there are expensive resources and conditions for manufacturing valuable products. Previously reports have shown specific microorganisms that can decompose keratinous waste. Further advancements to improve keratinase generation and its thorough characterization are required for the future application of keratinase in the context of decomposing keratin-incorporating waste on a mass scale.
In the present study, 20 actinomycetes were isolated from soils of Jeddah in Saudi Arabia. Among the total number of samples, AM8 was identified as the top candidate for feather decomposition. Feather decomposition initiated by bacteria has been actively studied for Bacillus spp., which is an omnipresent bacterium in feather and poultry waste.31 Many studies have been conducted on the isolation, growth dynamics, and keratinolytic capacities of Bacillus spp. isolated from wastewater from a poultry plant.32 Active actinobacterial isolates generating keratinase have been screened and quantitatively tested in a basal salt medium incorporating feathers as a single carbon.33 An improved approach for the characterization of keratinase activity and performance has been formulated for Alternaria tenussima and Aspergillus nidulans, which have been isolated from poultry waste.34 Importantly, keratinolytic performance has been measured by placing Bacillus licheniformis enzyme under incubation conditions inside the keratin solution.21 Biochemical and microscopic analyses were performed specifically on the AM8 isolate. Sangali and Brandelli35 conducted biochemical analyses to define Vibrio sp. strain kr 2. The present study identified AM8 actinobacteria based on morphological, physiological, and biochemical experiments and reports.36 Living colonies of S. coelicolor strain A3 growing on cellophane films were monitored using phase-contrast microscopy.37 The biochemical, physiological, and morphological characteristics of the strains observed were found to be dependent on their belonging to the genus Streptomyces. Evidently, the organism creates a clear phylogenetic line inside the 16S rRNA that is attributed to Streptomyces.38,39 The strain was noted as S. rochei AM8 for its effective application in the future. The enzymatic decomposition of chicken feathers results in an excessive amount of free amino acids, which may be further used for industrial purposes. Based on these results, there are preconditions to assert whether actinobacteria-generating keratinase may be utilized effectively in the industrial field.
The current research focused on studying 20 cultures of actinomycetes capable of producing keratinase. The cultures were isolated from sites where keratin-incorporating substrates exist in a natural setting. The AM8 isolate was identified and described based on its morphology, growth dynamics, and biochemical characteristics. It is associated with the genus Streptomyces. S. rochei AM8 exhibited the highest keratinase production potential (0.451 U/mL) among the isolates. The optimal production temperature was 35°C, whereas the optimal pH for the isolate was 7. Feather decomposition (92%) using isolated S. rochei AM8 has the potential to be used to effectively decompose feather waste under more coordinated and optimized conditions. The ability of the isolated bacteria to decompose feather waste is an opportunity to effectively manage feather waste from the poultry industry with the help of biotechnological resources. However, the study of the biotechnological use of keratinase requires more extensive insights, specifically about the triggers that provoke this enzyme to decompose native substrates of keratinase. Thus, further research is required to examine methods to purify keratinase. Moreover, information is needed on the dynamics and kinetics of enzymes, substrate ranges, the impact of inhibitors and triggers on enzyme performance, and submerged-state fermentation for the large-scale manufacturing of keratinase. Future research should examine whether a cohort of actinomycetes can be used for feather waste decomposition instead of relying on individual cultures to ensure improved keratinolytic potential.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in this manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants, or animals performed by any of the authors.
- Prasanna IL, Moorthy K. Isolation, Identification and Characterization of Feather Degrading Raoultella electrica from Poultry Waste Containing Soil. 2020.
- Khalel A, Alshehri W, Aly M. Enhancing Plant Growth by Chicken Feather Compost Obtained from Feather Degradation by Streptomyces enissocaesilis. Biosc Biotech Res Comm. 2020;13(4).
Crossref - Manirujjaman M, Amin R. Nahid AA, Alam S. Isolation and characterization of feather degrading bacteria from poultry waste. African Journal of Bacteriology Research. 2016;8(3):14-21.
- Korkmaz H, Hur H, Dincer S. Characterization of alkaline keratinase of Bacillus licheniformis strain HK-1 from poultry waste. Ann Microbiol. 2004;54(2):201-211
- Ramnani P, Singh R, Gupta R. Keratinolytic potential of Bacillus licheniformis rg1: structural and biochemical mechanism of feather degradation. Can J Microbiol. 2005;51(3):191-196.
Crossref - Nam GW, Lee DW, Lee HS, et al. Native feather degradation by Fervidobacterium islandicum AW-1, a newly isolated keratinase-producing thermophilic anaerobe. Arch Microbiol. 2002;178(6):538-547.
Crossref - Nagal S, Jain PC. Production of feather hydrolysate by Elizabethkingia meningoseptica KB042 (MTCC 8360) in submerged fermentation. Indian J Microbiol. 2010;50(Suppl 1):S41-S45.
Crossref - Sharma R, Gupta R. Thermostable, Thiol activated keratinase from Pseudomonas aeruginosa – KS1 for prospective applications in Prion decontamination. Res J Microbiol. 2010;5(10):954-965.
Crossref - Mabrouk MEM. Feather degradation by a new keratinolytic Streptomyces sp. MS- 2. World J Microbiol Biotechnol. 2008;24(10):2331-2338.
Crossref - Syed DG, Lee JC, Li WJ, Kim CJ, Agasar D. Production, Characterization and application of keratinase from Streptomyces gulbargensis. Bioresource Technol. 2009;100(5):1868-1871.
Crossref - Jani SA, Soni R, Patel H, Prajapati B, Patel G. Screening, isolation and characterization of keratin degrading actinomycetes: Streptomyces sp. and Saccharothrix xinjiangensi and analyzing their significance for production of keratinolytic protease and feed grade amino acids. Int J Curr Microbiol App Sci. 2014;3(9):940-955.
- Maruthi YA, Lakshmi A, Rao R, Chaitanya DA. Chrysosporium tropicum – A potential feather/hair waste degrading keratinophilic fungi. J Environ Res Manage. 2011;2:014- 018.
- Cao L, Tan H, Liu Y, Xue X, Zhou S. Characterization of a new keratinolytic Trichoderma atroviride strain F6 that completely degrades native chicken feather. The Society for Applied Microbiology. Lett Appl Microbiol. 2008;46(3):389-394.
Crossref - Gradisar H, Friedrich J, Krizaj I, Jerala R. Similarities and specificities of fungal keratinolytic proteases: comparison of keratinases of Paecilomyces marquandiiand Doratomyces microsporus to some known proteases. Appl Environ Microbiol. 2005;71(7):3420-3426.
Crossref - Anbu P, Gopinath SCB, Hilda A, Lakshmipriya A, Annadurai G. Purification of keratinase from poultry farm isolate Scopulariopsis brevicaulis and statistical optimization of enzyme activity. Enzyme Microb Technol. 2005;36(5-6):639-647.
Crossref - Rodrigues NM, Ledesma CT, Cirena DV, et al. New feather – degrading filamentous fungi. Microb Ecol. 2008;56(1):13-17.
Crossref - Anitha TS, Palanivelu P. Production and characterization of keratinolytic protease(s)from the fungus, Aspergillus parasiticus. International Journal of Research in Biological Sciences. 2012;2(2):87-93.
- Jain N, Sharma M. Biodiversity of Keratinophilic Fungal Flora in University Campus, Jaipur, India. Iranian J Publ Health. 2012;41(11):27-33.
- Kumawat TK, Sharma V, Seth R, Sharma A. Diversity of Keratin Degrading FungalFlora in Industrial area of Jaipur and Keratinolytic Potential of Trichophyton Mentagrophytes and MicrosporumCanis. International Journal of Biotechnology and Bioengineering Research. 2013;4(4):359- 364.
- Tamilmani P, Umamaheswari A, Vinayagam A, Prakash B. Production of extracellular feather degrading enzyms by Bacillus licheniformis isolated from poultry farm soil in Namakal Districk (Tamilnadu). Int J Poult Sci. 2008;7(2):184-188.
Crossref - Vigneshwaran C, Shanmuga S, Kumar TS. Screening and characterization of keratinase from Bacillus licheniformis isolated from Namakkal poultry farm. 2010;2(4):89-96.
- Matikeviciene V, Masiliuniene D, Grigiskis S. Degradation of keratin containing waste by Bacteria with keratinolytic activity”, Environmental Technology Resources Proceeding of the 7th International Scientific and Practical Conference. 2009;1.
Crossref - Suntornsuk W, Suntornsuk L. Feather degradation by Bacillus sp. FK 46 in submerged Cultivation. Bioresour Technol. 2003;86(3):239-243.
Crossref - Sastry TP, Sehgal PK, Gupta B, Kumar M. Solublised keratins as Novel filler in the retaining of upper leather. Leather Science. 1986;33:345-359.
- Macedo AJ, da Silva WOB, Gava R, Driemeier D, Henriques JAP, Termignoni C. Novel keratinase from Bacillus subtilis S14 exhibiting remarkable dehairing capabilities. Appl Environ Microbiol. 2005;71(1):594-596.
Crossref - Tork SE, Aly MM, Al-Fattani SQ. A new uricase from Bacillus cereus SKIII: Characterization, gene identification and genetic improvement. Int J Biol Macromol. 2020a;165(Part B):3135-3144.
Crossref - Lacey J. Actinomycetales: Characteristics and Practical mportance. Edited by G. Sykes and F. Skinner. The Society for Applied Bacteriology Symposium Series. Academic Press London- New York. 1973:2. https://www.worldcat.org/title/actinomycetales-characteristics-and-practical-importance/oclc/661590
- Letourneau F, Soussotte V, Bressollier P, Branland P,. Keratinolytic activity of Streptomyces sp. SK1-02: a new isolated strain. Lett Appl Microb. 1998;26(1):77-80.
Crossref - Williams ST, Vickers JC. Detection of actinomycetes in the natural environment-problems and perspectives. In Okami Y, Beppu T and Ogawara H. (eds.). Biology of actinomycetes ’88. Japan Scientific Societies Press, Tokyo. 1988:165-270. http://www.bioline.org.br/request?ac93008
- Goodfellow M, Lechevalier MP. Genus Nocardia. Trevisan. 9AL. In: Bergey’s Manual of Systematic Bacteriology. Edited by ST Williams ME, Sharpe JG. Holt. Baltimore: Williams & Wilkins. 1989;2:1458-1471. https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=43354
- Thys RCS, Lucas FS, Riffel A, Heeb P, Brandelli A. Characterization of a protease of a feather degrading Mycobacterium sp. Lett Appl Microb. 2004;39:181-186.
Crossref - Matikeviciene V, Masiliuniene D, Grigiskis S. Degradation of keratin containing wastes by bacteria with keratinolytic activity. Environ Technol Resour. 2009;1:284-289.
Crossref - Kawato M, Shinobu R. Coverslip culture of Streptomyces hertaricolor nov sp. Supplement; a simple technique for the microscopic observation. Mem Osaka Univ Lib Art Educ. 1959;8:114-119.
- Aneja K. Experiments in Microbiology Plant pathology and biotechnology. Forth Edition New age international limited publishers New Delhi. 2003.
- Sangali S, Brandelli A. Feather keratin hydrolysis by Vibrio sp. Strain kr2. J Appl Microbiol. 2000;89(5):735-743.
Crossref - Arifuzzaman M, Khatun MR, Rahman H. Isolation and screening of actiomycetes from sundarbans soil for antibacterial activity. Afr J Biotechnol. 2010;9:4615-4619.
- Donlan. Advance in applied microbiology. Book Reference. J. Read 30. 1983.
- Lapage SP, Sneath PHA, Lesse EF, Skerman VBD, Seeliger HPR, Clark WA. International code of nomenclature of Bacteria. American Society for Microbiology, Washington. 1976.
- Kuznetsov VD, Rodionova EG, Yangulova IV, Dmitrieva SV. Review of the systematic position of Actinomyces erythraeus Waksman et Curtis 1916 and its transfer to the genus Proactinomyces. Mikrobiologiya. 1977;46:114-118.
- Singhal S, Singhal N, Agarwal S. Pharmaceutical Analysis II, Thin layer chromatography, Pragati prakashan, First edition, 2009:98-111. http://ijpsr.com/bft-article/an-overview-on-thin-layer-chromatography/?view=fulltext
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.