ISSN: 0973-7510
E-ISSN: 2581-690X
Recently, the search for novel products derived from natural sources has become necessary due to the decreasing effectiveness of current antibiotics in treating bacteria that are antibiotic-resistant. In this context, it is well known that entomopathogenic bacteria (EPBs) produce a broad range of secondary metabolites with antibacterial activities. Therefore, an in-vitro trial was conducted to isolate and identify non-symbiotic bacteria associated with entomopathogenic nematodes, Steinernema spp. and evaluate the antibacterial activity against four antibiotic-resistant bacteria. Four bacterial isolates, i.e. Pseudomonas alcaligenes ST-1, Paenibacillus barcinonensis ST-2, Bacillus mojavensis ST-3, and Bacillus megaterium ST-4 were non-symbiotically isolated from the heamolymph of dead Steinernema-infected Galleria mellonella larvae and molecularly characterized. The bacterial cells and filtrates from P. alcaligenes ST-1 strongly inhibited the growth of Staphylococcus aureus through disk diffusion (43 mm), minimum inhibitory concentration (2.5 µL/mL), and minimal bactericidal concentration (5 µL/mL) assay. Conclusively, the direct application of endogenous Steinernema-associated EPB as an antibacterial agent for antibiotic-resistant bacteria looked promising.
Non-symbiotic Entomopathogenic Bacteria, Molecular Identification, Antibiotic-resistant Bacteria, Antibacterial Activity
Antibiotic resistance is currently one of the most serious worldwide health challenges. The abuse of antibiotics in both medicine and agriculture in industrialized and developing countries frequently results in antibiotic resistance mechanisms, which threaten modern medicine by decreasing the potency of therapeutically active drugs.1-3 Antibiotic-resistant bacteria including Staphylococcus aureus, Enterobacter cloacae, Klebsiella pneumoniae, Escherichia coli, Pseudomonas aeruginosa, Acinetobacter baumannii, Enterococcus faecalis, and Bacillus cereus have become an emerging public health problem.4 Bacteria have been exposed to long-term selective pressure due to the overuse of antibiotics, which has led to antibiotic resistance, making the bacteria difficult to eradicate. As a result, the expense of healthcare increases, illnesses last longer, more expensive drugs are used, and preventive and treatment are more effective. To effectively treat infections, antibiotics are utilized as a broad antimicrobial therapy. There are currently fewer antibiotics available that can prevent drug resistance, putting us at risk of running out of effective antibiotics in the future.2-5 Antibiotic-resistant bacteria, which are anticipated to cause roughly 10 million annual deaths by 2050, are difficult to quantify accurately.6 Therefore, antibiotic-resistant bacteria require alternative therapies. One such alternate strategy involves biological molecules derived from bacterial or natural sources. Gram-negative bacteria belonging to the Enterobacteriaceae family, Xenorhabdus and Photorhabdus are symbiotically associated with the infected juveniles (IJs) of entomopathogenic nematodes (EPNs) belonging to the Steinernema and Heterorhabditis genera, respectively.7 During IJ penetration, the non-symbiotic bacteria, Pseudomonas, Stenotrophomonas, Alcaligenes, Achromobacter, Pseudochrobactrum, Ochrobactrum, Brevundimonas, Deftia, etc. are spontaneously “hitchhiked” into IJ vectors through the cuticle or intercuticular space and injected into the insect hemocoel.8 It has been shown that bacterial resources for the synthesis of antimicrobial substances are either symbiotic or non-symbiotic bacteria associated with EPNs.8,9 Their cell and filtrate activities successfully inhibit the growth of S. aureus and S. pyogenes,10,11 Botrytis cinerea and Bacillus subtilis,12 K. pneumoniae, E. coli and E. coloacae,13 Fusicladium effusum,14 Phytophthora capsici, Bacillus anthracis and Rhizoctonia solani.15 These bacteria generate a wide spectrum of secondary metabolites, some of which have cytotoxic, insecticidal, and antibacterial properties.11,16-18 The following substances have been shown to be antimicrobial: 3,5-dihydroxy-4-isopropystilbene,12 1-carbapen- 2-em-3-carboxylic acid,13 benzaldehyde,15 3-hydroxy-2-isopropyl-5-phenethylphenyl carbamate,19 2-isopropyl-5-(3-phenyl-2-oxiranyl0-benzene-1,3diol,20 and chaiyaphumine.21 However, there is limited information on the antibiotic activity of the bacteria associated with EPNs from southwestern Saudi Arabia. This study’s goals were to isolate and identify non-symbiotic EPBs associated with Steinernema spp.; analyze their phylogenetic variability; and test the antibacterial potency of the identified strains against four antibi-otic-resistant bacteria: S. aureus, E. cloacae, E. coli, and B. cereus using the method of disk diffusion, minimum inhibitory concentration (MIC), and minimal bactericidal concentration (MBC).
Isolation of Steinernema-associated bacteria
EPNs, Steinernema spp. were previously isolated and morphologically identified22 by Prof. Dr. Ahmed Noureldeen, Biology Departement, Faculty of Sciences, Taif University, Taif, Saudi Arabia. Using the method described by Vitta et al.,23 non-symbiotic bacteria were isolated from the haemolymph of dead Galleria mellonella larvae that had been infected with the IJs of Steinernema spp.23 In Brief, the dead G. mellonella larvae were surface-sterilized by washing in 100% ethanol for 1 min, and then they were deposited in a sterile Petri dish to dry. After that, a sterile-sharp needle was used to cut the third segment from the larvae’s head, allowing a flow of symbiotic or non-symbiotic bacteria-containing haemolymph to emerge. The haemolymph samples were distributed and streaked over NBTA media (nutrient agar with 0.004% triphenyl tetrazolium chloride and 0.025% bromothymol blue) using a sterile loop, and then incubated at 28°C for 48 h.24 Regularly, bacteria were cultured every 24 h until they produced pure, isolated colonies, after which they were refrigerated for subsequent work at -80°C with 20% glycerol (v/v). A single colony of each bacterial isolate was seeded in 5 mL of Luria-Bertani (LB) broth and cultured with agitation at 220 rpm overnight at 28°C to create the cell-free conditioned filtrates or cell suspensions. Furthermore, 100-mL aliquots of culture were shaken at room temperature for 24 h before being placed to flasks containing 400 mL of the same media and shaken for five days at 200 rpm. The supernatant and bacterial pellets were obtained after centrifuging the multiplied bacterial culture (13,000 rpm for 30 min) at 4°C. In order to get a cell-free filtrate, the supernatant was filtered using a 0.22 μm Millipore filter, and the pellet was then resuspended in sterile distilled water. In addition to obtain concentrations of 100, 80, 60, 40, 20, 10, 5, and 2.5 μL/mL, the filtrate was further diluted with sterile distilled water and kept at 4°C. Using a spectrophotometer, the bacterial cell suspension was adjusted at OD600 to 1.0. The bacterial suspension had a concentration of 1×107 (CFU/mL) after using a 10-fold serial dilution spread plate.
Identification of Steinernema-associated bacteria
DNA extraction from bacterial isolates
Using the Bacterial Genomic DNA Miniprep Kit, the genomic DNA of the isolated bacteria was obtained from the bacterial pellets (QIAprep Spin Miniprep Kit). Prior to use in a PCR, the bacterial genomic DNA was kept in a fridge at -20°C.
PCR Amplification of 16S rRNA Gene
In the PCR reactions, universal oligonucleotide primers were used: 27F (5′-AGAGTTTGATCCTGGCTCAG-3′), and 1492R (5′ TACGGT TACCTTGTTACGACTT-3′).25 The final reaction volume was 50 μL and contained 1 μL of bacterial DNA, 22 μL of ddH2O, and 1 μL of each primer. In accordance with the manufacturer instruction, this mixture was added to the GoTaq® Green Master Mix (Promega, Madison, WI, USA). Then, carry out the following procedure using a C1000 TouchTM Thermal Cycler (Bio-Rad, Munich, Germany): A thermocycler was employed to complete 36 cycles, each consisting of three steps: denaturation (95°C for 1 min), annealing (54°C for 1 min), extension (72°C for 3 min), and final extension (72°C for 7 min).26 Conventional electrophoresis was used to analyze amplified products and identify bands. By using universal primer, the amplified products’ sizes were 1465 bp which indicates that the band is the 16S rRNA gene.
Sequencing of 16S rRNA gene
For four isolates that were packed in accordance with the National Health Research Ethics Committee’s approval and in accordance with the sequencing company’s guidelines, DNA purification and Standard Sanger sequencing were carried out (Macrogen Inc. Seoul, Korea). Under accession numbers (OP860651 to OP860654) on NCBI’s website (https://www.ncbi.nlm.nih.gov), the following sequences have been added.
Bioinformatic analysis
The Finch TV program was used to visualize nucleotide sequence isolates (version 1.4.0). The web program nucleotide BLAST (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) was used to explore Genbank databases for nucleotide sequence similarity.27 The BioEdit program (version 7.2) was used to investigate closely related sequences from NCBI and subject them to multiple sequence alignment.28 MEGA software was utilized to create the Neighbour-Joining phylogenetic tree with default settings (http://www.megasoftware.net/index.html).29
Antibiotic-resistant bacteria preparation
For antibacterial activity test, four isolates of antibiotic-resistant bacteria, Staphylococcus aureus, Enterobacter cloacae, Escherichia coli, and Bacillus cereus were employed. The Mueller-Hinton agar (MHA) was streaked with these bacteria, and it was then incubated at 37°C for 24 h. The turbidity was adjusted to 0.5 McFarland standards after dissolving a single colony in 0.85% sodium chloride (NaCl). For the disk diffusion test, a sample of the bacterial suspension in the amount of 100 μL was spread on MHA.30
Antibacterial activity
To initially evaluate the antibacterial activity of the four non-symbiotic bacterial cell suspensions or filtrates against antibiotic-resistant bacteria, 20 μL of cell suspension (107 CFU/mL) or cell-free supernatant (200 μL/mL) from each isolate was dropped on a Mueller-Hinton (MH) agar plate containing bacteria resistant to antibiotics. After that, the plates were incubated for 24 h at 37°C. Positive results were read in a clean zone from the margin of a non-symbiotic bacterial growing colony. For the disc sensitivity test, EPB isolates that potentially inhibited at least one antibiotic-resistant bacterium were chosen.
Disk diffusion method
Ten microliters of each filtrate concentration at 200 μL/mL or cell suspension concentration at 107 CFU/mL from the four bacterial isolates were poured into sterile 6 mm paper discs, which were subsequently placed on the MHA plated with antibiotic-resistant isolates. Ampicillin-containing antibiotic disc was served as a positive control, while distilled water-containing disc was employed as a negative control. After that, the plates were incubated for 24 h at 37°C. A ruler was used to measure the clear zone’s diameter (millimeter), which reflects the zone of inhibition.
Minimum Inhibitory Concentrations (MIC) and Minimal Bactericidal Concentrations (MBC)
Bacterial filtrates (Cell-free conditioned media) with the best disk diffusion results were subsequently assessed using the broth microdilution method to determine minimum inhibitory concentrations (MIC). In a 96-well micro titer plate, eight serial dilutions of the bacterial filtrate (100, 80, 60, 40, 20, 10, 5, and 2.5 μL/mL) were carried out. As a control, antibiotic-resistant bacteria were maintained in sterile Mueller-Hinton (MH) broth. For 24 h., plates were then incubated at 37°C. The MIC is the lowest concentration of filtrate at which the bacteria do not appear to be growing in the well. The minimal bactericidal concentrations (MBC) were also assessed. 96-well MIC micro titer plates with ten microliters from each well were sub-cultured onto MHA plates. The plates were then incubated for 24 h at 37°C. Each filtrate’s lowest concentration at which bacteria did not grow was classified as MBC. MIC and MBC assay were tested thrice.
Statistical analysis
The results were presented as mean ± standard error (M ± SE). Using the COSTAT program, the data were statistically evaluated by one-way analysis of variance (ANOVA), followed by multiple comparison, and significance was determined by P-values less than 0.05.
Phylogenetic analysis of 16S rRNA gene Sequences
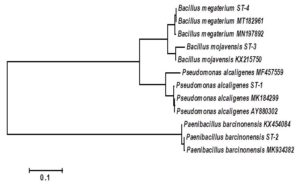
The accession numbers of the four isolated bacteria non-symbiotically associated with Steinernema spp. were OP860651– OP860654 as appeared in Table 1. The number of base pairs in the 16S rDNA sequences of the bacterial isolates studied varied significantly and identified as Pseudomonas alcaligenes, Paenibacillus barcinonensis, Bacillus mojavensis and Bacillus megaterium. The different bacterial isolates showed homology in the nucleotide sequence from 98 to 100%. To elucidate the genetic relatedness between the bacterial isolates and the related sequence of strains obtained from the NCBI database on the basis of the sequencing analysis of the 16S rDNA gene, a phylogenetic tree was constructed. The phylogeny tree based on the obtained 16S rDNA sequences was divided into two main clusters (Figure 1). The first cluster was divided into two subclusters. The first one contained Bacillus megaterium ST-4 which was closed similar to Bacillus megaterium MT182961 and Bacillus megaterium MN197892 with a similarity ratio of 100 and 98 %, respectively. On the same subcluster Bacillus mojavensis ST-3 was found closed similarity to Bacillus mojavensis KX215750 with about a similarity ratio of 98%. Moreover, the second subcluster contained Pseudomonas alcaligenes ST-1 which was closed similar to Pseudomonas alcaligenes MK184299 and Pseudomonas alcaligenes MF457559 with a similarity ratio of 99 and 98 %, respectively. On the other hand, the second main cluster was contained Paenibacillus barcinonensis ST-2 which was closed similar to Paenibacillus barcinonensis MK934382 and Paenibacillus barcinonensis KX454084 with a similarity ratio of 99 and 98 %, respectively.
Table (1):
The NCBI BLAST query for bacterial isolates.
Isolates |
Species |
Query coverage % |
E value |
Ident % |
Accession number |
|---|---|---|---|---|---|
ST-1 |
Pseudomonas alcaligenes |
100.00 |
0.00 |
99.00 |
OP860651 |
ST-2 |
Paenibacillus barcinonensis |
100.00 |
0.00 |
100.00 |
OP860652 |
ST-3 |
Bacillus mojavensis |
99.00 |
0.00 |
98.00 |
OP860653 |
ST-4 |
Bacillus megaterium |
100.00 |
0.00 |
99.00 |
OP860654 |
Figure 1. Neighbour-Joining phylogeny based on 16S rRNA gene sequences of non-symbiotic bacterial isolates related to Steinernema spp.
Bioactivity
Screening of EPB isolates against antibiotic-resistant bacteria
The data presented in Table 2 show that the whole cell culture and the filtrates of Steinerne-ma-associated bacteria had inhibition effect on tested antibiotic-resistant bacteria which varied from weak to strong inhibition. The antibiotic-resistant bacteria S. aureus, E. cloacae, and E. coli were susceptible to three of the EPB isolates, whereas, B. cereus was sensitive to two of the bacterial isolates. Among all tested bacterial isolates, the strongest inhibition activities have been recorded by P. alcaligenes ST-1 and B. megaterium ST-4 cells and filtrates against S. aureus, while only cells of those bacteria have the same effects on E. coli. However, cells and filtrates of P. alcaligenes ST-1 reported moderate inhibition on E. cloacae growth, whereas, B. megaterium ST-4 cells and filtrates gave weak inhibition on the same bacterium growth. No inhibition activities have been noticed when E. coli and B. cereus bacteria exposed to P. barcinonensis ST-2 cells or filtrates, while all of the antibiotic-resistant bacteria except E. coli had resistance to B. mojavensis ST-3
(Table 2).
Table (2):
Antibacterial activity of four bacterial species cells or filtrates on antibiotic-resistant bacteria
| Bacterial species | Bacterial suspension type | Inhibit the growth of antibiotic-resistant bacteria a | |||
|---|---|---|---|---|---|
| S. aureus | E. cloacae | E. coli | B. cereus | ||
| Pseudomonas alcaligenes ST-1 | Cells | +++ | ++ | +++ | ++ |
| Filtrates | +++ | ++ | ++ | + | |
| Paenibacillus barcinonensis ST-2 | Cells | ++ | + | – | – |
| Filtrates | + | + | – | – | |
| Bacillus mojavensis ST-3 | Cells | – | – | + | – |
| Filtrates | – | – | + | – | |
| Bacillus megaterium ST-4 | Cells | +++ | + | +++ | ++ |
| Filtrates | +++ | + | ++ | + | |
a No inhibition (-), weak inhibition (+), moderate inhibition (++), strong inhibition (+++).
Antibacterial activity using disk diffusion method
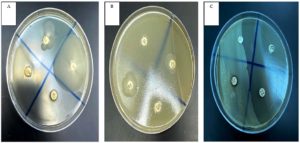
The antibacterial activities of either cells or filtrates of the four isolated non-symbiotic bacteria associated with Steinernema spp. were confirmed by disk diffusion method (Table 3, Figure 2). P. alcaligenes ST-1, P. barcinonensis ST-2, B. mojavensis ST-3, and B. megaterium ST-4 bacterial cells at 107 CFU/mL and cell free supernatant at 200 μL/mL caused inhibition zone (means ± SE) ranging from 14.4 to 43.0 mm, 0.0 to 16.6 mm, 0.0 to 10.5 mm, and 9.8 to 38.1 mm, respectively (P < 0.05). Bacterial cells of the four isolated strains were more effective against all of examined antibiotic-resistant bacteria than bacterial filtrates, inducing a mean clear zone of 12.96 mm for the cells and 10.21 mm for the filtrates (P < 0.05). The largest inhibition zone (43.0 mm) was significantly recorded when S. aureus bacteria were exposed to P. alcaligenes ST-1 cells, while it inhibited the growth of E. cloacae, E. coli, and B. cereus with the clear zone averaged 24.0, 31.3, and 22.8 mm, respectively. However, the cell free supernatant of P. alcaligenes ST-1 could inhibit the growth of S. aureus, E. cloacae, E. coli, and B. cereus with the clear zone 36.1, 20.5, 26.6, and 14.4 mm, respectively. Furthermore, the cells and filtrates of B. megaterium ST-4 were the second most toxic to the examined bacteria with the inhibition zone of 38.1 and 32.5 mm, 13.1 and 9.8 mm, 30.3 and 25.0 mm, and 20.1 and 13.5 mm for S. aureus, E. cloacae, E. coli, and B. cereus, respectively. In contrast, the smallest inhibition zone was recorded from E. coli (6.4 mm) treated with filtrates of B. mojavensis ST-3. Moreover, no inhibition activity was recorded from B. mojavensis ST-3 against S. aureus, E. cloacae, and B. cereus, and P. barcinonensis ST-2 against E. coli, and B. cereus (Table 3).
Table (3):
Antibacterial activity of four bacterial species cells or filtrates on antibiotic-resistant bacteria as assessed by disk diffusion.
| Bacterial species | Bacterial suspension type | Inhibition Zone (mm) a | |||
|---|---|---|---|---|---|
| S. aureus | E. cloacae | E. coli | B. cereus | ||
| Pseudomonas alcaligenes ST-1 | Cells | 43.0±2.07 | 24.0±1.51 | 31.3±1.67 | 22.8±1.67 |
| Filtrates | 36.1±2.42 | 20.5±1.77 | 26.6±1.41 | 14.4±0.74 | |
| Paenibacillus barcinonensis ST-2 | Cells | 16.6±0.74 | 9.3±1.03 | 0.0±0.0 | 0.0±0.0 |
| Filtrates | 13.1±1.25 | 6.3±1.03 | 0.0±0.0 | 0.0±0.0 | |
| Bacillus mojavensis ST-3 | Cells | 0.0±0.0 | 0.0±0.0 | 10.5±1.2 | 0.0±0.0 |
| Filtrates | 0.0±0.0 | 0.0±0.0 | 6.4±1.41 | 0.0±0.0 | |
| Bacillus megaterium ST-4 | Cells | 38.1±1.49 | 13.1±1.13 | 30.3±1.04 | 20.1±1.25 |
| Filtrates | 32.5±1.41 | 9.8±1.04 | 25.0±1.20 | 13.5±0.93 | |
| Ampicillin | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | |
a Four repetitions, each with two plates, were used in this experiment to represent each treatment. Numbers in each column indicated to inhibition zone diameter ± standard error.
Figure 2. Disk diffusion method of bacterial cell-free supernatant of non-symbiotic EPBs against antibiotic-resistant bacterium S. aureus. Clear zone of S. aureus after exposure to bacterial filtrate from P. alcaligenes ST-1 (A14), B. megaterium ST-4 (B17), P. barcinonensis ST-2 (B19), B. mojavensis ST-3 (B20), antibiotic disc (C21) and negative control (C22)
Minimum Inhibitory Concentrations (MIC)
The MIC values of the four isolated non-symbiotic bacteria associated with EPN Steinernema spp. were also determined against four bacterial strains as indicated in Table 4. EPB filtrates with MICs ranging from 2.5 to 80 µL/mL inhibited the growth of antibiotic-resistant bacteria. The microorganism that recorded highest sensitivity towards P. alcaligenes ST-1 filtrate was S. aureus (2.5 µL/mL), followed by E. coli (5 µL/mL), then E. cloacae (20 µL/mL), and B. cereus (40 µL/mL). All of the testes bacteria were susceptible to B. megaterium ST-4 filtrate, which had the strongest effects (10 µL/mL) against S. aureus and E. coli., followed by E. cloacae (40 µL/mL), and B. cereus (60 µL/mL), respectively. P. barcinonensis ST-2 presented activity against S. aureus (60 µL/mL) and E. cloacae (80 µL/mL), however, B. mojavensis ST-3 bacterial filtrate was active only against E. coli with the MIC value (80 µL/mL).
Table (4):
Minimum inhibitory concentration of bacterial filtrates (µL/mL) against antibiotic-resistant bacteria.
| Bacterial species | MIC (µL/mL) a | |||
|---|---|---|---|---|
| S. aureus | E. cloacae | E. coli | B. cereus | |
| Pseudomonas alcaligenes ST-1 | 2.5 | 20 | 5 | 40 |
| Paenibacillus barcinonensis ST-2 | 60 | 80 | ND | ND |
| Bacillus mojavensis ST-3 | ND | ND | 80 | ND |
| Bacillus megaterium ST-4 | 10 | 40 | 10 | 60 |
a Each treatment in this experiment had three repetitions. Numbers in each column indicated to minimum inhibitory concentration of each non-symbiotic bacterium. ND= Not detected.
Minimal Bactericidal Concentrations (MBC)
Likewise, it seemed that the effective MIC also corresponds to the bacteria’s effective bactericidal concentration (Table 5). The antibiotic-resistant bacterium S. aureus was the most susceptible to all filtrates except B. mojavensis ST-3 with MBCs ranging from 5 to 60 µL/mL. The activity of P. alcaligenes ST-1 filtrate against S. aureus was lower than that of all other tested bacteria with the MBC (5 µL/mL), followed by E. coli (10 µL/mL), while it recorded 40 and 60 µL/mL for E. cloacae, and B. cereus, respectively. E. cloaca was the least sensitive bacteria suppressing by P. barcinonensis ST-2 and scored highest MBC value (100 µL/mL).
Table (5):
Minimal bactericidal concentrations of EPB filtrates (µL/mL) against antibiotic-resistant bacteria.
| Bacterial species | MBC (µL/mL) a | |||
|---|---|---|---|---|
| S. aureus | E. cloacae | E. coli | B. cereus | |
| Pseudomonas alcaligenes ST-1 | 5 | 40 | 10 | 60 |
| Paenibacillus barcinonensis ST-2 | 60 | 100 | ND | ND |
| Bacillus mojavensis ST-3 | ND | ND | 80 | ND |
| Bacillus megaterium ST-4 | 10 | 60 | 20 | 80 |
a Each treatment in this experiment was represented by three replicates. Numbers in each column indicated to minimal bactericidal concentration of each non-symbiotic bacterium. ND= Not detected.
This study’s early stage was successful in recovering and isolating non-symbiotic EPB from Steinernema spp. Four Steinernema-associated bacteria were identified. These results are in line with those found by,9 who isolated five EPBs associated with Steinernema feltiae and evaluated their potentials against two aphid insects, Aphis punicae and Aphis illinoisensis. Accordingly, my investigations confirmed the previous results of,31 who isolated in the same region Photorhabdus sp. and Xenorhabdus sp. from the Heterorhabditis sp. and Steinernema sp., respectively, and documented their complex activities against the root-knot nematode, Meloidogyne incognita, which infects pomegranate under greenhouse conditions. The present study’s findings definitely demonstrated that bacterial culture and 16S rDNA sequence analysis offer a novel method for comparing different bacterial community patterns. Based on a phylogenetic tree analysis, the four isolated species of Steinernema-associated bacteria discovered here were named separately and molecularly identified as P. alcaligenes ST-1, P. barcinonensis ST-2, B. mojavensis ST-3, and B. megaterium ST-4. These findings agreed with those of other research.32-34,25 Data had a high bootstrap percentage and demonstrated how the most isolated strains related to one another. The other isolates, in contrast, had longer branch lengths, which were in line with earlier discoveries, and this was explained by the incomplete 16S rDNA sequences.34-36 In the present study, the four isolated bacteria from the heamolymph of insect G. mellonella were non-symbiotically related to Steinernema spp. infection, and have been first identified in Saudi Arabia. These outcomes are consistent with those that8 have previously recorded, on four species of Steinernema and their associated bacteria. Regarding the antibacterial activity, it was evident that the four bacterial isolates cells were stronger than bacterial filtrates in their ability to combat antibiotic-resistant bacteria, which showed low toxicity. The different isolates of EPBs have been reported with variation in antibacterial activity. This might be as a result of each bacterium’s ability to synthesize therapeutic metabolites or the sensitivity of bacteria that are resistant to antibiotics. Among the examined bacterial isolates, P. alcaligenes ST-1 and B. megaterium ST-4 cells and filtrates exhibited the strongest inhibitory effect, whereas, B. mojavensis ST-3 displayed the weakest inhibition activity. These results are in consent with those of37 who noted that high diversity of antimicrobial components present in B. megaterium strains make them prospective sources of novel antibiotics produced by bacilli and demonstrate efficacy against test strains that are resistant to pharmaceutical antibiotics (gram-negative bacteria P. aeruginosa ATCC 27853 and E. coli ATCC 25922, glycopeptide-resistant leuconostoc, and methicillin-resistant staphylococcus). These findings are also in agreement with those of38,39 who mentioned that b-Sitosterol, behenic acid, phenylacetic acid, and other identified Bacillus megaterium L2 compounds show promise as potential natural antimicrobial agents by destroying cell membrane integrity, damaging cell structures, affecting cell metabolism, and inhibiting protein synthesis to exhibit an antibacterial effect. Similarly, a novel cyclic peptide compound from B. megaterium was quantified and showed broad spectrum antimicrobial activity against both gram positive and gram negative bacteria, with minimum inhibitory concentrations of 6.25, 3.125, 1.0, 0.5, and 0.25 µg/mL against S. aureus ATCC25923, P. aeruginosa ATCC27853, E. coli ATCC35218, Salmonella typhi ATCC19430, and Micrococcus luteus ATCC10240, respectively, with no activity against C. albicans ATCC10231.40 The obtained data also revealed that S. aureus and E. cloacae were sensitive to all isolated bacterial cells and filtrates except B. mojavensis ST-3; however, E. coli was susceptible to all isolates except P. barcinonensis ST-2; meanwhile, B. cereus showed sensitivity to P. alcaligenes ST-1 and B. megaterium ST-4 only. Accordingly, based on mean inhibition zone values, the toxicity caused by studied EPBs against tested antibiotic-resistant bacteria could be categorized in the descending order: P. alcaligenes ST-1 > B. megaterium ST-4 > P. barcinonensis ST-2 > B. mojavensis ST-3; whereas, the sensitivity of the antibiotic-resistant bacteria could be arranged as S. aureus > E. coli > E. cloacae > B. cereus. These results are in the same context with the previous studies regarding the EPBs, Photorahbdus and Xenorhabdus those showed that P. luminescens could inhibit the growth of drug-resistant and clinical isolates B. subtilis, E. coli, and S. aureus RN4220.13 Additionally, S. aureus ATCC20475, S. aureus strain PB36, and S. aureus strain PB57 were also vulnerable to inhibitory activity of Photorhabdus extracts.41 Likewise, Xenorhabdus synthesized xenocoumacin42 and amicoumacin43 derivatives, both of which were discovered to be effective antibiotics against S. aureus.10 However, isopropylstilbene,44,45 which all Photorhabdus spp. produced exhibited a wide range of biological properties including antibiotic action against S. aureus and E. coli.19 The results showed that the EPBs P. barcinonensis ST-2 and B. mojavensis ST-3 have less bactericidal potential against antibiotic-resistant bacteria based on the MIC and MBC. In contrast, earlier investigations show that B. mojavensis, and its secreted compounds, lipopeptides, and fengycin effectively suppress a number of bacterial and fungal strains.46,47 Similarly, Singh et al.48 screened the inhibitory activity of Paenibacilllus sp. and found that S. aureus (ATCC 25923), E. coli (MTCC 1610), C. albicans (MTCC 224), and Bacillus subtilis (ATCC 6633), were all inhibited by the filtrate and/or crude fermentation extract of fifteen isolates, suggesting the antibacterial activity due to compound bacitracin A. Also, peptides isolated and purified from Paenibacillus peoriae IBSD35 cell-free supernatant showed a high and broad spectrum of antimicrobial activity.49 Here, the author reported that the non-symbionts EPBs were isolated for the first time from Steinernema-infected G. mellonella heamolymph and considered as antibacterial agents. Of these, the symbiont, Xenorhabdus was not recorded in current study. This might be as a result of the bacterium’s limited distribution. This is consistent with the findings of earlier research in Taif region, Saudi Arabia which recorded the nematicidal and insecticidal9,31,22 activities. Overall, the obtained results revealed that different species of antibiotic-resistant bacteria were affected differently by EPB isolates. This might be caused by each bacterium’s ability to generate valuable metabolites or by antibiotic-resistant bacteria’s sensitivity to the specific metabolites manufactured by each non-symbiont.
In the current study, four bacterial isolates were non-symbiotically associated with Steinernema spp., and be identified as the first recorded at Taif, Saudi Arabia. Their antibacterial efficacy was evaluated against four important antibiotic-resistant bacteria. P. alcaligenes ST-1 amongst acts as an excellent antibacterial agent to the four tested antibiotic-resistant bacteria. The findings provide a reliable base for the discovery of a novel bioactive compound.
ACKNOWLEDGMENTS
The author would like to thank Dr. Ahmed Noureldeen for providing the EPN.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in this manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by the author.
- Yalew ST. Review on antibiotic resistance: resistance mechanisms, methods of detection and its con-trolling strategies. Biomed J Sci Tech Res. 2020;24(5):18651-18657.
Crossref - Siedlecka A. The occurrence of antibiotic resistance genes in tap water-a review. EPJ Web of Conferences. 2018;30:9.
Crossref - Vicar EK, Acquah SEK, Williams W, Kuugbee ED, Saba CKS, Mensah GI. Antibiotic resistant bacteria infecting wounds of rural community dwellers in northern Ghana. Eur J Med Health Sci. 2021;3(1):112-117.
Crossref - Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis. 2018;197(8):1079-1081.
Crossref - Roe VA. Antibiotic resistance: a guide for effective prescribing in women’s health. J Midwifery Women’s Health. 2018;53(3):216-226.
Crossref - Chokshi A, Sifri Z, Cennimo D, Horng H. Global Contributors to Antibiotic Resistance. J Glob Infect Dis. 2019;11(1):36-42.
Crossref - Boemare NE, Akhurst RJ, Mourant RG. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int J Syst Bacteriol. 1993;43(2):249-255.
Crossref - Ogier JC, Pages S, Frayssinet M, Gaudriault S. Entomopathogenic nematode-associated microbiota: From monoxenic par-adigm to pathobiome. Microbiome. 2020;8:25.
Crossref - Baazeem A, Alotaibi SS, Khalaf LK, et al. Identification and environment-friendly biocontrol potential of five dif-ferent bacteria against Aphis punicae and Aphis illinoisensis (Hemiptera: Aphididae). Front Microbiol. 2022;13:961349.
Crossref - McInerney BV, Taylor WC, Lacey MJ, Akhurst RJ, Gregson RP. Biologically active metabolites from Xenorhabdus spp., Part 2. Benzopyran-1-one derivatives with gastroprotective activity. J Nat Prod. 1991;54(3):785-795.
Crossref - Bode HB. Entomopathogenic bacteria as a source of secondary metabolites. Curr Opin Chem Biol. 2009;13(2):224-230.
Crossref - Chen G. Antimicrobial Activity of the Nematode Symbionts, Xenorhabdus and Photorhabdus (En-tero-bacteriaceae), and the Discovery of Two Groups of Antimicrobial Substances, Nematophin and Xenorxides. In: Canada (Doctoral dissertation). British Columbia: Simon Fraser University, 1996.
- Derzelle S, Duchaud E, Kunst F, Danchin A, Bertin P. Identification, Characterization, and Regulation of a Cluster of Genes Involved in Carbapenem Biosynthesis in Photorhabdus luminescens. Appl Environ Microbiol. 2002;68(8):3780-3789.
Crossref - Bock CH, Shapiro-Ilan DI, Wedge DE, Cantrell CL. Identification of the antifungal compound, trans-cinnamic acid, produced by Photorhabdus luminescens, potential biopesticide against pecan scab. J Pest Sci. 2014;87:155-162.
Crossref - Ullah I, Khan AL, Ali L, et al. Benzaldehyde as an insecticidal, antimi-crobial, and antioxidant compound produced by Photorhabdus temperata M1021. J Microbiol. 2015;53(2):127-133.
Crossref - Chacon-Orozco JG, Bueno Jr C, Shapiro-Ilan DI, Hazir S, Leite LG, Harakava R. Antifungal activity of Xenorhabdus spp. and Photorhabdus spp. against the soybean pathogenic Sclerotinia sclerotiorum. Sci Rep. 2020;10(1):1-12.
Crossref - Yuksel E, Ozdemir E, Delialioglu RA, Canhilal R. Insecticidal activities of the local entomopathogenic nema-todes and cell-free supernatants from their symbiotic bacteria against the larvae of fall webworm, Hyphantria cunea. Exp Parasitol. 2022;242:108380.
Crossref - Fodor A, Clarke DJ, Dillman AR, Tarasco E, Hazir S. New Antimicrobial Peptides from Bacte-ria/Invertebrate Obligate Symbiotic Associations. Front Microbiol. 2022;13:862198.
Crossref - Shi D, An R, Zhang W, Zhang G, Yu Z. Stilbene derivatives from Photorhabdus temperata SN259 and their antifungal activities against phytopathogenic fungi. J Agric Food Chem. 2017;65(1):60-65.
Crossref - Hu KJ, Li JX, Li B, Webster JM, Chen G. A novel antimicrobial epoxide isolated from larval Galleria mellonella infected by the nematode symbiont, Photorhabdus luminescens (Enterobacteriaceae). Bioorg Med Chem. 2006;14(13):4677-468.
Crossref - Grundmann F, Kaiser M, Schiell M, et al. Antiparasitic Chaiyaphumines from entomopathogenic Xenorhabdus sp. PB61.4. J Nat Prod. 2014;77(4):779-783.
Crossref - Alotaibi SS, Darwish H, Zaynab M, et al. Isolation, Identification, and Biocontrol Potential of Entomopathogenic Nematodes and Associated Bacteria against Virachola livia (Lepidoptera: Lycaenidae) and Ectomyelois ceratoniae (Lepidoptera: Pyralidae). Biology. 2022;11(1):295.
Crossref - Vitta A, Thimpoo P, Meesil W, et al. Larvicidal activity of Xenorhabdus and Photorhabdus bacteria against Aedes aegypti and Aedes albopictus. Asian Pac J Trop Biomed. 2018;8(1):31-36.
Crossref - Yooyangket T, Muangpat P, Polseela R, Tandhavanant S, Thanwisai A, Vitta A. Identification of en-to-mopathogenic nematodes and symbiotic bacteria from Nam Nao National Park in Thailand and larvicidal activity of symbiotic bacteria against Aedes aegypti and Aedes albopictus. PLoS ONE. 2018;13(4):e0195681.
Crossref - Liu W, Sun Z, Zhang J, et al. Analysis of microbial composition in acid whey for dairy fan making in Yunnan by conventional method and 16S rRNA se-quencing. Curr Microbiol. 2009;59(2):199-205.
Crossref - Alsanie WF, Felemban EM, Farid MA, Hassan MM, Sabry A, Gaber A. Molecular Identification and Phylo-genetic Analysis of Multidrug-resistant Bacteria using 16S rDNA Sequencing. J Pure Appl Microbiol. 2018;12(2):489-496.
Crossref - Hassan MM, Farid MA, Gaber A. Rapid identification of Trichoderma koningiopsis and Trichoderma longibrachiatum using sequence characterized amplified region markers. Egyp J Biol Pest Con. 2019;29:13.
Crossref - Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947-2948.
Crossref - Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870-1874.
Crossref - Seier-Petersen MA, Jasni A, Aarestrup FM, et al. Effect of subinhibitory concentrations of four commonly used biocides on the conjugative transfer of Tn916 in Bacillus subtilis. J Antimicrob Chemother. 2014;69(2):343-348.
Crossref - Nour El-Deen AH, Al-Barty AF, Darwesh HY, Al-Ghamdi AS. Eco-Friendly Management of Root-Knot Nem-atode, Meloidogyne incognita Infecting Pomegranate at Taif Governorate, KSA. Res J Pharm Biol Chem Sci. 2016;7(1):1070-1076.
- Clarridge JE. Impact of 16S rDNA Gene Sequence Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases. Clin Microbiol Rev. 2004;17(4):840-862.
Crossref - Anand P, Chellaram TC, Kumarn S, Felicia SC. Screening for antibiotic producing marine bacterial against fish pathogens. Int J Pharma Bio Sci. 2011;2(1):314-325.
- Hassan MM, Ismail AI. Isolation and molecular characterization of some pathogenic mobile phone bac-teria. Int J Biochem Biotechnol. 2014;3(3):516-522.
- Sabir JMS, Abo-Aba SEM, Sabry A, Hussein RM, Bahieldin A, Baeshen NA. Isolation, identification and comparative analysis of 16S rDNA of Bacillus subtilis grown around Rhazya stricta roots. Life Science Journal. 2013;10(12s):980- 986.
- Alfatih YM, Idris AB, Hassan HG, et al. Detection of a novel mutation G511T in the 530 loop in 16S rRNA in multi drugs resistant Mycobacterium tuberculosis isolated from Sudanese patients. BioRxiv. 2018;1:497628.
Crossref - Malanicheva IA, Kozlov DG, Sumarukova IG, et al. Antimicrobial Activity of Bacillus megaterium Strains. Mikrobiologiya. 2012;81(2):196-204.
Crossref - Xie YD, Peng QJ, Ji YY, et al. Isolation and Identification of Antibacterial Bioactive Compounds from Bacillus megaterium L2. Front Microbiol. 2021;12:645484.
Crossref - Pan H, Xiao Y, Xie A, et al. The antibacterial mechanism of phe-nylacetic acid isolated from Bacillus megaterium L2 against Agrobacterium tumefaciens. Peer J. 2022;10:e14304.
Crossref - Al-Thubiani ASA, Yahia AM, Adel F, et al. Iden-tification and characterization of a novel antimicrobial peptide compound produced by Bacillus megaterium strain isolated from oral microflora. Saudi Pharm J. 2018;26(8):1089-1097.
Crossref - Muangpat P, Yooyangket T, Fukruksa C, et al. Screening of the Anti-microbial Activity against Drug Resistant Bacteria of Photorhabdus and Xenorhabdus Associated with Entomopathogenic Nematodes from Mae Wong National Park, Thailand. Front Microbiol. 2017;8:1142.
Crossref - Reimer D, Luxenburger E, Brachmann AO, Bode HB. A new type of pyrrolidine biosynthesis is involved in the late steps of Xenocoumacin production in Xenorhabdus nematophila. Chem Bio Chem. 2009;10(12):1997-2001.
Crossref - Park H, Perez C, Perry E, Crawford JM. Activating and attenuating the amicoumacin antibiotics. Molecules. 2016;21(7):E824.
Crossref - Li J, Chen G, Wu H, Webster JM. Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens. Appl Environ Microbiol. 1995;61(12):4329-4333.
Crossref - Buscato EL, Buttner D, Bruggerhoff KFM, et al. From a multipotent stilbene to soluble epoxide hydrolase inhibitors with antiproliferative properties. Chem Med Chem. 2013;8(6):919-923.
Crossref - Ghazala I, Charfeddine S, Charfeddine M, Gargouri R, Ellouze S, Haddar A. Antimicrobial Activities and An-tioxidant Properties of Bacillus Mojavensis I4 Lipopeptides and Application in the Biocontrol of Potato Dry Rots Caused by Fusarium Solani. Arch Microbiol. 2022.
Crossref - Jasim B, Sreelakshmi S, Mathew J, Radhakrishnan EK. Identification of endophytic Bacillus mojavensis with highly specialized broad spectrum antibacterial activity. 3 Biotech. 2016;6:187.
Crossref - Singh H, Manpreet K, Manoj J, Sunita M, Hemraj N, Anil KP. Antimicrobial properties of the novel bacterial isolate Paenibacilllus sp. SMB1 from a halo-alkaline lake in India. Sci Rep. 2019;9:11561.
Crossref - Ngashangva N, Pulok K, Mukherjee CS, Mohan CK, Indira S. Integrated genomics and proteomics analysis of Paenibacillus peoriae IBSD35 and insights into its antimicrobial characteristics. Sci Rep. 2022;12:18861.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.