ISSN: 0973-7510
E-ISSN: 2581-690X
Bioluminescence, the capacity of living creatures to produce light through chemical processes, has been a topic of scientific fascination for many years. Bioluminescence is of great importance in the marine environment, since it facilitates diverse ecological interactions. Bioluminescent bacteria establish symbiotic partnerships with marine creatures, such as squids, octopus, dinoflagellets, various fishes like anglerfishes, etc. In order to use bioluminescent bacteria as a biosensor, the samples of Indian Squid were gathered from local fish market located in Ganesh Peth and Camp area of Pune district, Maharashtra. This study was performed to isolate and characterize potential bioluminescent bacteria which can be used as biosensors in food and brewing industry for food analysis. In this study, 608 colonies with blue color and 873 colonies with green color bioluminescence were isolated from Indian squid using sea water agar. The bacterial strains AB28 and ABG67 were selected for further analysis, which required comparing them with the information provided in Bergerys handbook for their characterization. Additionally, their identification was carried out using 16S rRNA. The bioluminescent strains that were isolated and identified were Shewanella seohaensis and Shewanella hafniensis, respectively.
AB28, ABG67, Bioluminescence, Indian Squid, Shewanella, Sea Water Agar
Bioluminescence is the emission of light that occurs in living organisms as a result of an oxidation reaction catalyzed by enzymes. Thus, the spectrum range of light emission emitted by bioluminescent organisms is around 400-700 nm, or blue-red light. The most prevalent colors in light emission are blue and green color variation.1 The affinity for this color is due to the preferred habitat of bioluminescent creatures, primarily found in seawater, which is most effectively penetrated by blue light.1-3 The relevant genes for this phenomenon are collectively known as the lux operon, which has luciferase enzymes.2 Bioluminescence is initiated by the oxidation of a long-chain aliphatic aldehyde and the reduction of flavin mononucleotide (FMNH2). This process requires oxygen and is facilitated by the enzyme luciferase.4
FMNH2 + RCHO + O2 → FMN + RCOOH + H2O + light5
The genes luxICDABEG are essentially included in the lux operon. The LuxA and LuxB genes encode for the a and b subunits of the luciferase enzyme, respectively. However, lux CDE genes are responsible for encoding polypeptides known as fatty acid reductase genes.5 Squid utilize the luminescence generated by bacteria to engage in an activity referred to as counterillumination.6 Luminous bacteria have been discovered in three families: Vibrionaceae, Shewanellaceae, and Enterobacteriaceae. These bacteria belong to five primary genera: Vibrio, Photobacterium, Aliivibrio, Photorhabdus and Shewanella. Bioluminescent bacteria exhibit a high degree of sensitivity towards a wide range of hazardous or toxic substances. Consequently, efforts have been undertaken to utilize this characteristic in many areas of environmental monitoring by employing either naturally occurring strains or by creating genetically modified bioluminescent bacteria.3,7-15 This study was carried out to isolate and characterize potential bioluminescent bacteria which can be utilized for food analysis.
Sample collection
In this study, the collection of samples and isolation procedure was carried out in two seasons summer (March) and winter (November). The environmental temperature in the summer was between 14 °C to 39 °C while in winter it was between 8 °C to 28 °C. Indian squid was collected from local fish market (Day 0) in Ganesh peth and near Camp area, Pune and stored in freezer for 15 days for enrichment of microorganisms.7 Different fish parts including eye part, tentacles, arms, body surface and abdomen and ink sac after dissection were used for isolation of bioluminescent bacteria.7
Evaluation of different media used for bioluminescent bacteria and isolation of bioluminescent bacteria
Freshly obtained fish was used for isolation of bacteria as well as fish after enrichment for 15 days (D15) were used. Swabs from different body parts as well as after dissection, under sterile conditions were taken and streaked on various media like Photobacterium Agar (PBA),16 Luminous Agar (LA),16,17 Zobell marine agar,2 Sea Water Agar (SWA: g/L: Part A: yeast extract 5.0, peptic digest of animal tissue 5.0, beef extract 3.0, agar 15.0; Part B: NaCl 24.0, KCl 0.7, MgCl2·6H2O 5.3, MgSO4·7H2O 7.0, CaCl2 0.1, pH 7.5 ± 0.2),5,6 plates and incubated at room temperature,6,17,18 37 °C5 and 30 °C for 24 hrs After incubation period, plates were observed in dark room for presence of bioluminescence. Isolated bioluminescent bacteria were further subcultured 8-9 times to get pure culture and maintained on Sea Water Agar media (Himedia, India).
Morphological and biochemical characterization of bacteria
Bacteria were macroscopically characterized based on the color, size, and shape of their colonies, as well as their colony morphology, elevation, and form. The microscopic analysis involved techniques such as gram staining to identify the type of bacteria, including cocci, spiral, and rod-shaped bacteria. Additionally, the motility of the organisms was assessed using the conventional hanging drop method. Different biochemical methods were used for identification of bacteria such as methyl red, Indole production test, voges Proskauer test, Citrate utilization test, Glucose and sucrose fermentation test, Catalase test, oxidase test and nitrate reduction test were studied.5 Organisms were also screened for their growth on TCBS selective media.9 Bacteria were also screened for production of enzymes like amylase, cellulase, Lipase, gelatinase and casein hydrolysate.5 Luminescence spectra of these bacteria was analysed by fluorescence spectrophotometer19 and the relation between growth and luminescence intensity was studied. The growth of bacteria was studied by measuring the turbidity of the suspension after every hour at 600 nm using spectrophometer while its intensity was studied at LUX400 nm using fluorescence spectrophotometer and its correlation was studied statistically. Purified bacterial strains were cultivated in TYS broth. This broth was used for preparation of glycerol stocks.20 These glycerol stocks were stored at -20 °C for further studies.
16S rRNA sequence analysis and identification
The identification of isolates was carried out at the sequencing facility of National Centre for Microbial Resource (NCMR), National Centre for Cell Science, Pune. At the facility, genomic DNA was isolated by the standard phenol/chloroform extraction method,21 followed by PCR amplification of the 16S rRNA gene using universal primers 16F27 [5′-CCA GAG TTT GAT CMT GGC TCA G-3′] and 16R1492 [5′-TAC GGY TAC CTT GTT ACG ACT T-3′]. The amplified 16S rRNA gene PCR product was purified by PEG-NaCl precipitation and directly sequenced on an ABI® 3730XL automated DNA sequencer (Applied Biosystems, Inc., Foster City, CA) as per manufacturer’s instructions. Essentially, sequencing was carried out from both ends using additional internal primers so that each position was read at least twice. Assembly was carried out using Lasergene package followed by identification using the EzBioCloud database.22
The observation that seasons play a significant role in the isolation of bioluminescent bacteria, with winter being particularly favorable, raises intriguing questions about the environmental factors influencing these organisms. Bioluminescent bacteria are often associated with deep ocean environments, where temperatures are lower, potentially explaining why the winter season, characterized by colder temperatures, enhances their growth and isolation. This suggests a preference or adaptation of these organisms to colder conditions, possibly linked to their natural habitat within deep-sea organisms like squids. The enrichment of bioluminescent bacteria through freezing the squid (D15) highlights the importance of nutrient availability from the squid gut. Observing the sea water agar plates in dark revealed that the plates for eye part, tentacles, arms, body surface did not show any glow while abdomen and ink sac culture were glowing in dark. The squid’s abdomen and ink sac, where glowing cultures were observed, likely provide rich nutrient environments conducive to bacterial growth and bioluminescence. This finding underscores the symbiotic relationship between the squid and these bacteria, where nutrient exchanges may support bacterial proliferation. Organisms from the abdomen displayed green bioluminescence, while those from the ink sac exhibited blue, reflecting potential ecological and biochemical adaptations. Blue light, with shorter wavelengths, penetrates seawater more effectively, making it suitable for long-distance communication or prey attraction in deep-sea environments In contrast, green light, with longer wavelengths, may serve better for short-range interactions, such as host-specific signaling or camouflage. The differentiation in luminescence colors likely reflects niche specialization. Green bioluminescence from the abdomen could aid the squid in mimicking ambient light patterns or signaling within close-range environments. Blue luminescence from the ink sac may play roles in predator distraction or prey attraction, particularly during defensive behaviors like ink release.
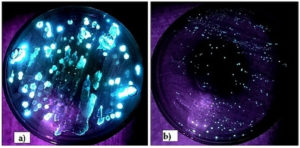
These variations are influenced by differences in luciferase enzymes and substrates, adapted to local environmental conditions such as pH, oxygen levels, or the host’s internal environment. This functional diversification not only reduces competition among bacterial strains but also highlights evolutionary pressures favoring specific luminescence types for ecological roles, ensuring survival and effective interaction within marine ecosystems. A total 608 isolates with blue color Bioluminescence were obtained while 873 colonies were showing green bioluminescence (Figure 1). Two Colonies showing maximum bioluminescence were selected by visual assessment from abdomen (AB28) (Figure 1a) and Ink sac (ABG67) (Figure 1b) were sub-cultured and used for further studies, while the D0 (Day 0) samples also showed bioluminescent bacteria in ink sac (7 isolates) and abdomen (32 isolates) plates but these organisms required regular subculture and lost their bioluminescence after 5 days and plates for eye part, tentacles, arms, body surface did not show any glow. The Photobacterium agar and luminous Agar showed growth of microorganisms but they did not glow in dark.
Figure 1. Bioluminescent colonies grown on Sea Water Agar: a) Isolates with blue colour luminescence from abdomen, b) Isolates with green colour luminescence from ink sac
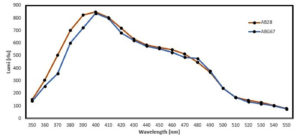
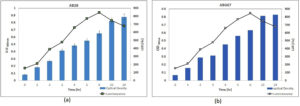
Morphological Characterization of AB28 and ABG67 was carried out. AB28 and ABG67 colonies were off white in color having puncti form and were circular, pin-point with convex elevation (Table). They were having entire margin. The AB28 was emitting Blue color Bioluminescence while ABG67 was emitting Green color luminescence in dark. Both isolates revealed typical features of gram-negative rods with motility, consistent with characteristics often found in marine bioluminescent bacteria. The motility observed, particularly in ABG67, suggests potential adaptations for movement within their environment, which could be advantageous for symbiotic relationships or dispersal in the squid’s body. Furthermore, biochemical tests indicating enzyme production profiles (e.g., catalase, oxidase, nitrate reductase, amylase, casein hydrolysis) provide insights into metabolic capabilities that support bacterial survival and function. The negative results for indole production and MRVP tests, coupled with positive fermentative abilities for glucose and sucrose, contribute to the microbial profile of these isolates, suggesting specialized adaptations possibly linked to their ecological niche. The luminescence spectra of isolated colonies suspensions were spectroscopically analyzed, revealing a luminescence peak at 400 nm (LUX400) with intensities around 800 relative luminescence units (rlu) (Figure 2). A positive correlation was observed statistically between the growth of bacteria and their intensity. As the bacteria number increased the turbidity also increase upto 7 hrs but after 23 hrs the intensity decreased and the turbidity remained constant indicating that the cells entered stationary phase. In exponential phase the cell density increases which correlates with the increase in turbidity and increase in intensity (Figure 3).
Table:
Characterization of the isolates AB 28 and ABG67
| Isolates | AB 281 Shewanella seohaenis | ABG673 Shewanella hafniensis |
|---|---|---|
| Morphological Characteristics | ||
| Colony color | Off white | Off white |
| Colony size | Pin-point | Pin-point |
| Colony shape | Circular | Circular |
| Colony elevation | Convex | Convex |
| Colony form | Punctiform | Punctiform |
| Margin | Entire | Entire |
| Light Emitted | Blue | Green |
| Gram staining | Gram-negative | Gram-negative |
| Bacteria Shape | Small rod | Small rod |
| Motility | Motile | Highly motile |
| Biochemical Characterization | ||
| MR | – | – |
| Indole | – | – |
| VP | – | – |
| Citrate test | + | + |
| Glucose | + | + |
| Sucrose | + | + |
| Catalase | + | + |
| Oxidase | + | + |
| Nitrate | + | + |
| TCBS | – | + |
| Enzyme production | ||
| Amylase | + | + |
| Cellulase | – | – |
| Gelatinase | – | – |
| Casein hydrolysate | + | + |
*Foot note: + = Positive, – = Negative
The genetic characterization through 16S rRNA sequencing (Figure 4), identifying AB28 and ABG67 as belonging to the genus Shewanella, highlights their phylogenetic relationship and potential evolutionary history. The high similarity with known species such as Shewanella seohaensis, Shewanella hafniensis highlight their classification within a recognized group of marine bacteria with Assession number GU944672 and AB205566 respectively, providing a basis for further genomic exploration and comparative studies.
This study reveals intriguing insights into the ecology and characteristics of bioluminescent bacteria associated with squids, particularly highlighting their preference for cold environments and symbiotic relationship with their host. The seasonal variation observed underscores the influence of environmental factors, with winter promoting the isolation and enrichment of these bacteria, likely due to the colder temperatures prevalent in deep-ocean habitats where squids reside. The distinct bioluminescence colors emitted from the abdomen (green) and ink sac (blue) suggest specialized adaptations and biochemical diversity among the isolated strains, potentially linked to their ecological roles within the squid. Morphological and biochemical characterizations confirm typical features of marine bioluminescent bacteria, with motility and specific enzymatic activities contributing to their ecological fitness. Genetic analysis places the isolates within the genus Shewanella, highlighting their phylogenetic affinity with recognized group of marine bacteria. This classification opens avenues for further genomic and evolutionary studies to elucidate their genetic adaptations and bioluminescence mechanisms. Overall, these findings contribute to our understanding of marine symbiosis and bioluminescence, providing a foundation for future research into the ecological significance and biotechnological potential of these fascinating microorganisms. Further exploration could focus on their interactions with squid hosts, mechanisms of bioluminescence regulation, and applications in environmental monitoring such as and biotechnology. These organisms can be further used to develop biosensor or bio-monitoring tools for commercial application in development of toxicity assays for water quality assessment and detection of food quality such as presence of heavy metals.
ACKNOWLEDGMENTS
The authors would like to acknowledge Dr. Jayant Londhe and Mrs. Suvarna Vaidya from Department of Biotechnology, Sinhgad College of Science and Dr. Rajashree Patwardhan from Department of Environmental Science and Microbiology, H.V. Desai College for providing necessary help throughout the study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SM conceptualized and designed the study. NK and ST performed data acquisition. NK, SM and ST performed data analysis. SM performed data interpretation. SM wrote the manuscript. SA and GP reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Brodl E, Winkler A, Macheroux P. Molecular Mechanisms of Bacterial Bioluminescence. Comput Struct Biotechnol J. 2018;16:551-564.
Crossref - BJ Bluth SEFBM. Cell-Cell Communication and the lux operon in Vibrio fischeri. https://www.bio.cmu.edu/courses/03441/TermPapers/97TermPapers/lux/default.html . Published online December 5, 2013.
- Widder EA. Bioluminescence in the ocean: origins of biological, chemical, and ecological diversity. Science. 2010;328(5979):704-708.
Crossref - Yaser NA, Abdullah MFF, Aris AM, Zainudin II. Isolation and Identification of Bioluminescent Bacteria in Squid and Water of Malaysia. International Journal of Advances in Agricultural and Environmental Engineering. 2014;1(2).
Crossref - Kannahi M, Sivasankari S. Isolation and Identification of Bioluminescent Bacteria from Marine Water at Nagapattinam Sea Shore Area. Int J Pharm Sci Rev Res. 2014;26(2):346-351.
- Kumar AR. Isolation of luminescent bacteria from bay of bengal and their molecular characterization. https://www.diva-portal.org/smash/get/diva2:1312151/FULLTEXT01.pdf
- Parmar P, Shukla A, Saraf M, Patel B. Isolation of Bioluminescent Bacteria from Marine Organisms. Indian Journal of Geo-Marine Sciences. 2020;49(03).
- Parmar P, Shukla A, Goswami D, Gaur S, Patel B, Saraf M. Comprehensive depiction of novel heavy metal tolerant and EPS producing bioluminescent Vibrio alginolyticus PBR1 and V. rotiferianus PBL1 confined from marine organisms. Microbiol Res. 2020;238:126526.
Crossref - Alloush HM, Lewis RJ, Salisbury VC. Bacteria bioluminescent biosensors: Food and Environment Monitoring. Analytical Letters. 2006;39:1517-1526.
Crossref - Griffiths MW. Applications of Bioluminescence in the Dairy Industry. J Dairy Sci. 1993;76:3118-3125.
Crossref - Syed AJ, Anderson JC. Applications of bioluminescence in biotechnology and beyond. Chem Soc Rev. 2021;50(9):5668-5705.
Crossref - Trif C, Vunduk J, Parcharoen Y, Bualuang A, Marks RS. Bioluminescent Whole-Cell Bioreporter Bacterial Panel for Sustainable Screening and Discovery of Bioactive Compounds Derived from Mushrooms. Biosensors. 2024;14(11):558.
Crossref - Hu L, Rossetti M, Bergua JF, et al. Harnessing Bioluminescent Bacteria to Develop an Enzymatic-free Enzyme-linked immunosorbent assay for the Detection of Clinically Relevant Biomarkers. ACS Appl Mater Interfaces. 2024;16(24):30636-30647.
Crossref - Zhang K, Liu M, Song X, Wang D. Application of Luminescent Bacteria Bioassay in the Detection of Pollutants in Soil. Sustainability (Switzerland). 2023;15(9):7351.
Crossref - Robinson GM, Tonks KM, Thorn RMS, Reynolds DM. Application of bacterial bioluminescence to assess the efficacy of fast-acting biocides. Antimicrob Agents Chemother. 2011;55(11):5214-5219.
Crossref - Bayyana S, Yalla SK, Cherian T, Mohanraju R. Isolation and characterization of bioluminescent bacteria associated with uroteuthis duvaucelli, an indian squid from andaman waters. Glbobal journal of bioscience and biotechnology. 2018;7(3):353-358.
- Kulkarni VS, Kulkarni BS. Isolation of Bioluminescent Bacteria and Their Application in Toxicity Testing of Chromium in Water. Int J Curr Microbiol App Sci. 2015;4(10):23-32
- Yaser NA, Abdullah MFF, Aris AM, Zainudin II. Isolation and Identification of bioluminescent bacteria in Squid and waters of Malyasia. Int’l Journal of Advances in Agricultural & Environmental Engg. (IJAAEE). 2014;1(2):EISSN 2349-1531.
- Marcelo GA, Galhano J, Duarte MP, Capelo-Martinez JL, Lodeiro C, Oliveira E. Validation of a Standard Luminescence Method for the Fast Determination of the Antimicrobial Activity of Nanoparticles in Escherichia coli. Nanomaterials. 2022;12(13):2164.
Crossref - Zhang XY, Han XX, Chen XL, et al. Diversity of cultivable protease-producing bacteria in sediments of Jiaozhou Bay, China. Front Microbiol. 2015;6:1021.
Crossref - Sambrook J, Fritsch ER, Maniatis T. Molecular Cloning: A Laboratory Manual (2nd ed.). New York: Cold Spring Harbor Laboratory Press., 1989.
- Yoon SH, Ha SM, Kwon S, et al. Introducing EzBioCloud: A Taxonomically United Database of 16S rRNA and Whole Genome Assemblies. Int J Syst Evol Microbiol. 2017;67(5):001755.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.