ISSN: 0973-7510
E-ISSN: 2581-690X
Heavy metals found in nature and the excessive accumulations of heavy metals have an impact on humans and animals. Different effects resulted from the toxicity of heavy metal, that damage the functioning of different organs: brain, lungs, kidney and other essential organs and lowering the levels of energy. Cadmium and lead considered being toxic to organisms in a particular concentration. Patients with renal failure concerned contaminated drinking water with cadmium and lead. Biofilm produced by microbes and it is important for the remediation of pollutants. This study aimed to isolate and investigate the ability of bacterial isolates to produce a biofilm that can resistance heavy metals (cadmium chloride (CdCl2) and lead nitrate Pb (No3)2). Isolates were isolated from soil sample located at different locations from Saudi Arabia (Makkah, Taif and Jeddah). Fifty isolates have been tested for formation of biofilm by two methods. First method was Congo Red Agar CRA and the second was Tissue Culture Plate TCP. Results revealed that 3 out of 50 isolates showed high biofilm formation. The three (A2, ST and PS) isolates that form strong biofilm were screened primarily on nutrient agar plate contain 7ppm concentration of CdCl2 and Pb (NO3)2. Results indicated that all three isolates were resistance. The maximum tolerance concentration (MTC) of three (A2, ST and PS) isolates studied on nutrient agar plate supplemented with different concentrations from CdCl2 and Pb (NO3)2 respectively. Results indicated that MTC values of Pb (NO3)2 were up to (450, 350 and 500 ppm) for ST, A2 and PS isolates respectively. While in CdCl2 the MTC values were (150, 120 and 250 ppm) for ST, A2 and PS isolates respectively. The effect of CdCl2 and Pb (NO3)2 on bacterial growth using spectrophotometer, and results indicated that all three isolates (A2, ST and PS) growth decreased with the increase in concentration of Pb (NO3)2 and CdCl2. Three isolates were identified by biochemical and 16S rRNA gene. The isolates identified as B. cereus A2, B. cereus ST and P.aeruginosa PS and submitted to NCBI under accretion numbers (MK450303 and MK450304 for B. cereus A2, B. cereus ST) respectively. Plasmid curing was studied using the method of elevated temperature, and results showed that all cured B. cereus A2 and P. aeruginosa PS colonies were resistance to 7ppm of CdCl2 and Pb (NO3)2 while B. cereus ST showed different pattern of resistance after curing. B. cereus ST selected for test removal Pb (NO3)2 and CdCl2 using inductively coupled plasma optical emission spectrometry (ICP-OES). Using ICPOES showed removal lead up to 93% while in cadmium to 49 %. Antimicrobial susceptibilities patterns of identified bacteria were determined. All tested isolated strains showed resistance against to 3 or more antibiotics. Three strains B. cereus A2, B. cereus ST and P. aeruginosa PS that isolated from soil, showed the highest biofilm formation which considered important factor for heavy metals resistance. The biofilm represents a very renewable, promising, cost-effective and easy biotechnology for treatment of wide range contaminated effluents.
Heavy metals, biofilm, genomic DNA, 16S rRNA sequencing, plasmid curing, B. cereus ST, lead nitrate Pb (NO3)2 Cadmium chloride (CdCl2), phylogenetic, Gram stain.
The heavy metals are non-degradable, toxic, tend to accumulate in organisms and undergo food chain amplification, persistent nature and effects on the local users, so considered the most dangerous groups (Rajeev Kumar et al., 2014). Badr et al., (2008) revealed that, the activities in the coastal area of Saudi Arabia by industries and human have increased through three decades, and resulted in heavy metals pollution. There are three main categories of metals: non-toxic and essential as Calcium (Ca) and Magnesium (Mg), only toxic at high concentrations (typically, Iron (Fe) Manganese (Mn), Zinc (Zn), Copper (Cu), Cobalt (Co), Nickel (Ni) and Molybdenum (Mo), and Mercury (Hg) or Cadmium (Cd) were toxic (Valls and de Lorenzo, 2002). Air, soil, and water are contaminated with toxic heavy metals that effect on human (McDonald and Grandt, 1981; Alloway, 1995).
Pandey and Jain, (2002) reported that the pollution with chemical and environmental problems has brought the possibility of long-term environmental disasters into the public conscience. Heavy metals including lead, mercury, cadmium and arsenic effects in human metabolism due to their persistence in the environment and documented potential for serious health consequences. Microorganisms can be used for the clean-up of environmental pollutants, this is known as bioremediation. Paul et al., (2005) reported that treating toxic effluents with the biological processes are the best efficiency and economy from other methods as (chemical and physical), and the potential of biofilm communities for bioremediation processes has been realized. Bioremediation with biofilm is safer because the cells in a biofilm possess good chance of adaptation and survival (during periods of stress), also as they are protected within the matrix so it is alternative to bioremediation with planktonic microorganisms (Decho, 2000). A biofilm includes one or more bacterial strains disposed in a matrix containing extracellular polymeric substances such as DNA, protein or carbohydrates (Post et al., 2013). Extracellular polymeric substances (EPS) are biosynthetic polymers produced by micro-organisms from (prokaryotic and eukaryotic) (Wingender et al., 1999).
Different factors effect on EPS production by bacterial strains in (culture or aggregates) depends on the microbial species, phases of growth, nutritional status and the environmental conditions (Sheng et al., 2006). Cell adhesion, formation of microbial aggregates (biofilms, flocs, sludges and bio-granules), depend on bacterial EPS (Sutherland, 2001; Tay et al., 2001; Comte et al., 2006) and also protect cells from hostile environments. EPS also involved in the degradation of particulate substances sorption of dissolved materials including heavy metals (Gutnick and Bach, 2000).
Different resistance mechanisms to counteract stress heavy metal have developed in bacteria, as the formation and sequestration of heavy metals in complexes, reduction of metal to a fewer toxic species, and direct efflux of a metal out of the cell (Nies, 1999), Environmentally significant microbe such as P.aeruginosa is a ubiquitous, possess many mechanisms of resistance, such as the mer operon that reduces toxic Hg2 to volatile HgO, which then diffuses out of the cell (Outten et al., 2000).
Morillo et al., (2006) reported that EPS produced from Paenibacillus jamilae strain capable of absorbing heavy metals from a multi-metal sorption system: Pb, Cd, Cu, Zn, Ni, Co when grown in aqueous extracts of two-phase olive mill waste.
Identification of bacteria from environmental or clinical specimens by sequencing of 16S rRNA gene that determine bacterial phylogenetic relationships (Vandamme et al., 1996; Gomila et al., 2004).
The present study investigated the (1) ability of some bacterial isolates that isolated from soils to produce a biofilm that can detoxify heavy metals (2) identified the bacterial strains by biochemical and 16S rRNA gene, (3) test removal Pb (NO3)2 and CdCl2, plasmid curing and antibiotic resistance.
Soil sample collection and bacterial isolations
All samples taken from the soil located at different locations from Saudi Arabia (Makkah, Taif and Jeddah). These samples were taken in sterilized polyethylene bags using the sterilized spatula and stored at 4°C until the examination. One gram of soil was suspended in 9 ml of sterilized distilled water, and serial dilutions up to 10 7 were prepared, 0.1ml suspension of each 10-5 and 10-7 dilutions spread on nutrient agar medium then plates incubated at 30°C for 24 hrs. Several colonies of bacteria were selected and isolated.
Biofilm Formation detection by Congo Red Agar method (CRA)
This medium prepared with brain heart infusion broth, sucrose, agar and Congo Red indicator. The stain of Congo Red was prepared separately as a concentrated aqueous solution and autoclaved. Then it was added to the autoclaved brain heart infusion agar with sucrose. Inoculate the plates of CRA with test bacterial isolates and incubated at 37oC for 24 hrs. Aerobically, black colonies indicated production biofilm (Freeman et al., 1989).
Biofilm Formation detection using Tissue Culture Plate method (TCP)
This assay is considered as a standard test for the detection the formation of biofilm. The overnight cultures grown in NB were diluted at 10-3 and inoculated into six individual wells of a Tissue Culture Plate Method (150ml per well). Then the plates were incubated for 24 hrs. at 30oC. Bacterial isolates were screened for their ability to form biofilm by the TCP method with a modification of (Christensen et al., 1985) according to (O’Toole and Kolter, 1998).
Heavy Metal Resistance Screening
The isolated bacterial colonies were screened for resistance to Cadmium Chloride (CdCl2) and Lead Nitrate (Pb (NO3)2 by adding 7ppm of CdCl2 and Pb (NO3)2, respectively, to sterilized nutrient agar medium. The isolated bacterial colonies were spot inoculated onto the plates of nutrient agar, then incubated for 24 hrs. at 37°C (Margeay et al., 1985).
Maximum Tolerable Concentration of Bacterial Isolates (MTC)
The maximum tolerable concentration is the highest concentration of the heavy metals that growth at 37°C. Isolated bacterial colonies that grew initially on the nutrient agar supplemented with the heavy metals CdCl2 and Pb (NO3)2 were exposed to increasing heavy metals concentrations (7ppm -500ppm). Testing for tolerance of the microorganisms ended when complete inhibition of the growth was observed on the nutrient agar with metal supplementation according to (Mulik and Bhadekar, 2017).
Growth Study of Metal Resistant Isolates
The capacity bacterial strains for tolerance of heavy metals were tested, cells were grown in 50 ml LB medium supplemented with different concentrations of the heavy metals, cadmium and lead, incubated for 24, 48 and 72 hrs. at 37°C on a rotary shaker (150 rpm) of selected heavy metal (cadmium (CdCl2), lead (Pb (NO3)2). Growth was monitored as a function of biomass by measuring absorbance at 600 nm using a spectrophotometer. The growth of the isolates on LB with no metal supplementation (control) and with metal supplementation (test) were performed and compared by plotting the optical density at 600 nm (OD600nm) to time in hours. According to (Chien et al., 2013).
Morphological Characterization and Biochemical Test
Bacterial isolates were tested for morphology on nutrient agar medium, microbiological tests such as Gram staining followed by biochemical identification tests like catalase, citrate oxidase, indole production.
Isolation of Genomic DNA
Bacterial colonies isolated from the nutrient agar with metal supplementations were characterized molecularly using the 16S rRNA sequencing. The genomic DNA was extracted using a GeneJET Genomic DNA extraction kit according to the manufacturer’s instructions.
Amplification of 16S rRNA
Amplification of 16S rRNA from extracted DNA has used as a template for PCR to amplify the 16S rRNA gene. The forward primer 27F 5′ (AGA GTT TGA TCM TGG CTC AG) 3 and reverse primer 1492R 5′(TAC GGY TAC CTT GTT ACG ACT T)3′ were used to amplify the 16s rRNA gene. PCR was performed with a one gel electrophoresis in 1 x TAE buffer with ethidium bromide (0.5 mg/ml) using the Mupid-One.
Sequencing of Amplified Fragments of Isolate 16S rRNA Genes
Samples for sequencing sent to MACROGEN, Korea. Sequences have compared with the available sequences against the 16S rRNA sequences database using NCBI’s BlastN. Sequences were aligned using the ClustalW program in Mega 6.0. Similarity index was generated and compared with known sequences.
Plasmid Curing using elevated Temperature for Bacterial Isolates
Ten ml of NB was inoculated by single colony, incubated for 24 hrs. at 37°C then 0.2 ml of bacterial culture was transferred to 10 ml of fresh NB and incubated at 45°C (an elevated temperature) for 24 hrs., with shaking at 100 rpm. Several dilutions up to 10-7 were prepared, then 0.1 ml of the last three dilutions were spread on plates of nutrient agar which supplemented with 7 ppm with CdCl2 and Pb (NO3)2 and incubated for 24 hrs. (37°C) (Kheder, 2002).
Gel Electrophoresis
According to (Sambrook et al, 1989), a plasmid was characterized by agarose gel electrophoresis through a gel of 1% agarose submerged in 100 ml 1X TBE supplemented with 2ml of ethidium bromide running buffer at 120 V for 1 hr. 7ml from sample with 1ml from loading buffer dye. DNA bands were visualized on UV. The plasmids MW was compared and determined by using DNA ladder.
Measurements of
CdCl2 and Pb (NO3)2 Removal by Bacterial Isolate
B. cereus ST, was grown in LB medium supplemented with different concentrations ranged from (7-150 ppm) of CdCl2 and (20-450 ppm) for Pb(NO3)2 individually. After 24, 48 and 72 hrs. of incubation, centrifuge at 10,000 rpm for 10 min. to cells separate the cells. The CdCl2 and Pb (NO3)2 removal properties were estimated by measuring metals depletion in culture supernatants by inductively coupled plasma-optical emission spectroscopy (ICP-OES). The ICP-OES system was calibrated by serial dilution of each metals standard (Mulik and Bhadekari, 2017).
Determination Bacterial Antibiotic Sensitivity and Resistance
Three bacterial strains on Mueller Hinton agar plates was tested against10 antibiotics: Tigecycline TGM (15mg/ml), Levofloxacinycin LEV (5mg/ml), Fusidic acid FA (10mg/ml), Vancomycin VA (30mg/ml), Taxo A (10mg/ml), Tobramycin TMN (10 mcg), Metronidazole MET (50 mcg), Ceftriaxone CRO (30mg/ml), Doxycycline DOX (30mg/ml), Aztreonam ATM (30mg/ml). Then inhibition zone diameters (IZD) were measured in mm. after 24 hrs. of incubation at 37°C. The strains were classified as being resistant, intermediate resistant or susceptible to a particular antibiotic. (-) Sensitive (S) ³ 21mm; intermediate (I) (16-20 mm) and resistant (R) £ 15mm (Thokchom and Joshi, 2012).
Samples collection and bacterial isolation
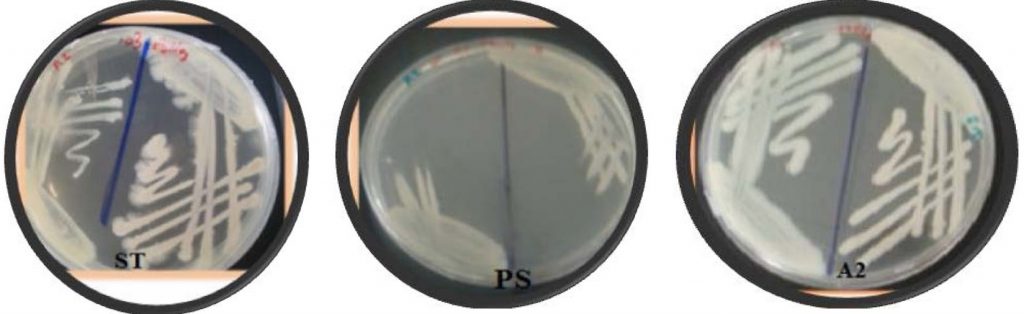
A total of fifty bacterial isolates were selected and storage at 4oC until test their ability for biofilm formation. The biofilm formation was verified using two techniques, one qualitative and the other quantitative. The qualitative technique of biofilm formation for fifty isolates was assessed by congo red agar. The black colors were observed for the biofilm production. Results indicated that several isolates were positive for biofilm formation. The results in (Fig. 1 and Fig. 2) showed the strong three isolates (A2, ST and PS) give positive black color colonies on Congo red agar plate while negative isolates give pink color indicating non-biofilm formation.
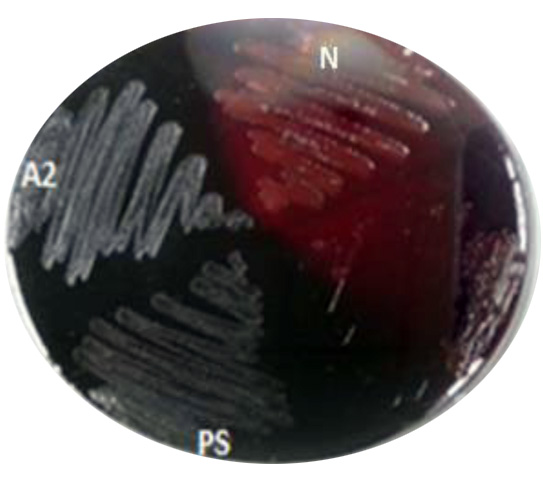
Fig. 2. Black color showing positive biofilm formation of A2 and PS isolate. Using CRA. While N: negative pink color for (non-biofilm formation)
Biofilm Formation Detection by Tissue Culture Plate Method (TCP)
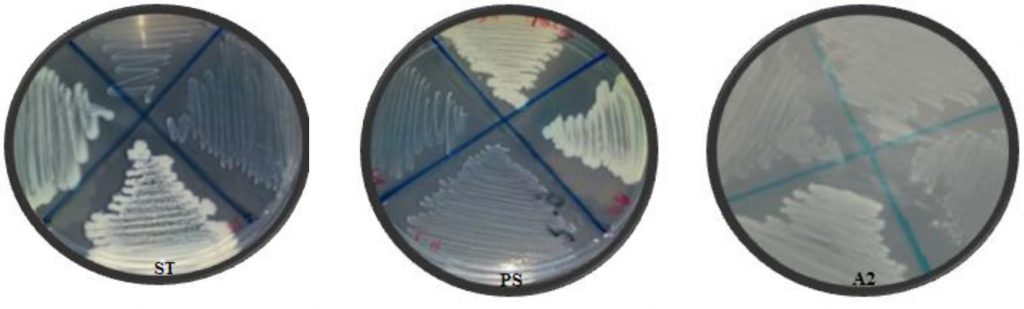
The second method for biofilm quantitative technique was Tissue Culture Plate (TCP). Fifty isolates were screened for their ability to form biofilm production by TCP were measure by using Micro-plate Reader at (OD570 nm) and considered zero (<0.05) biofilm formation, weak (0.05–0.12), moderate (0.12–0.24), or high (>0.24) according to TCP method (Gad et al., 2009). Results of biofilm production by TCP method revealed that 6% (3/50) were biofilm formation by Micro-plate Reader at optical density (OD570nm) as shown in (Table 1). Results indicated that isolates (ST, A2 and PS) OD570nm were (0.381, 0.380 and 0.436) respectively which considered high biofilm formation. The results showed that isolates (A2, ST and PS) were strong biofilm adherence (Fig. 3).
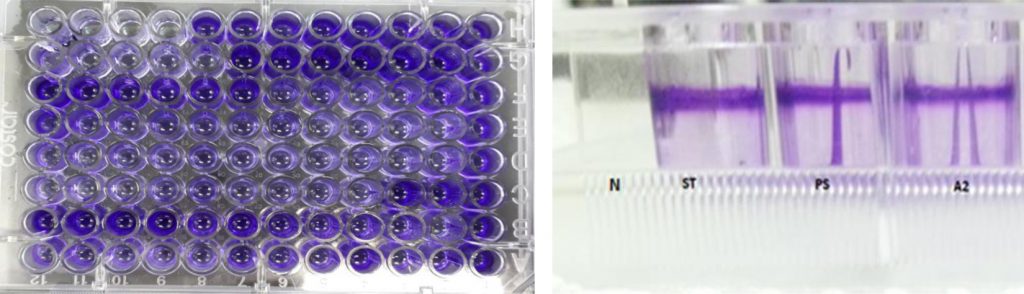
Fig. 3. Isolates (ST, A2 and PS) showed strong biofilm formation by TCP. While N showed non- biofilm formation. Screening for Heavy Metal Resistance
Screening of heavy metals Pb (No3)2 and CdCl2 resistance of the three isolates (A2, PS and ST) that form strong biofilm were tested on nutrient agar plate supplemented with 7 ppm of Pb (No3)2 and CdCl2 then incubated for 24 hrs. at 37°C Results showed that (A2, PS and ST) isolates were resistance to Pb(No3)2 and CdCl2.
Table (1):
Biofilm formation by micro-titer plate at OD570 nm for three bacterial isolates:
*Standard |
PS |
A2 |
ST |
Isolates |
|---|---|---|---|---|
0.24> |
0.436 |
0.380 |
0.381 |
(OD570 nm) |
High |
High |
High |
High |
Biofilm Formation |
Strong |
Strong |
Strong |
Strong |
Adherence |
*Micro-plate Reader at (OD570 nm) and considered zero (<0.05) biofilm formation, weak (0.05–0.12), moderate (0.12–0.24), or high (>0.24).
Maximum Tolerable Concentration (MTC)
(MTC) is the highest concentration of the heavy metal that allows growth after two days, isolates (A2, PS and ST) were tested. The results in (Table 2) indicated that ST isolate MTC tolerant up to 450 ppm of Pb(NO3)2, while A2 isolate MTC tolerant up to 350 ppm and PS isolate MTC tolerant up to 500 ppm. Results of CdCl2 showed in (Table 3) that ST, A2 and PS isolate MTC were ranged from (7-150 ppm) (7-120 ppm) and (7-250 ppm) respectively.
Table (2):
Maximum Tolerable Concentration of Lead Nitrate (ppm):
| Isolates | MTC of cadmium chloride(ppm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 10 | 20 | 50 | 100 | 120 | 150 | 200 | 250 | 300 | |
| ST | + | + | + | + | + | + | + | − | − | − |
| A2
|
+ | + | + | + | + | + | − | − | − | − |
| PS | + | + | + | + | + | + | + | + | + | − |
Table (3):
Maximum Tolerable Concentration of Cadmium Chloride (ppm)
| Isolates | MTC of Lead nitrate (ppm) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 20 | 50 | 100 | 150 | 200 | 250 | 300 | 350 | 400 | 450 | 500 | 550 | |||
| ST | + | + | + | + | + | + | + | + | + | + | + | − | − | ||
| A2 | + | + | + | + | + | + | + | + | + | − | − | − | − | ||
| PS | + | + | + | + | + | + | + | + | + | + | + | + | − | ||
Determination of the Effect of Pb (NO3)2 and CdCl2 on Bacterial Growth
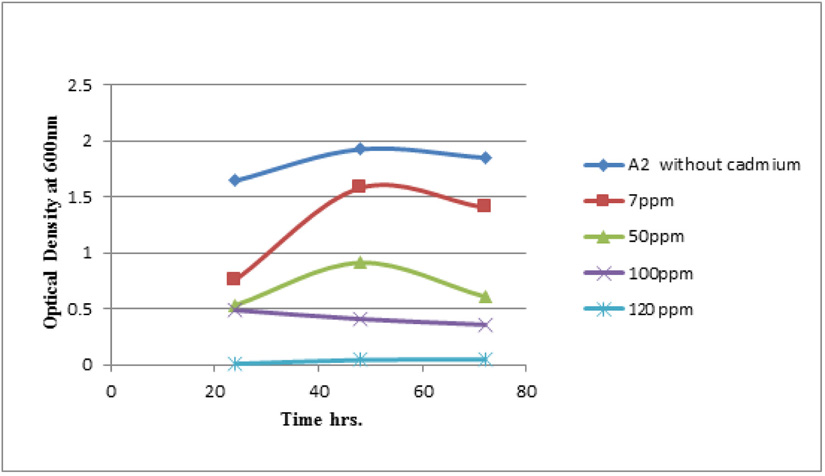
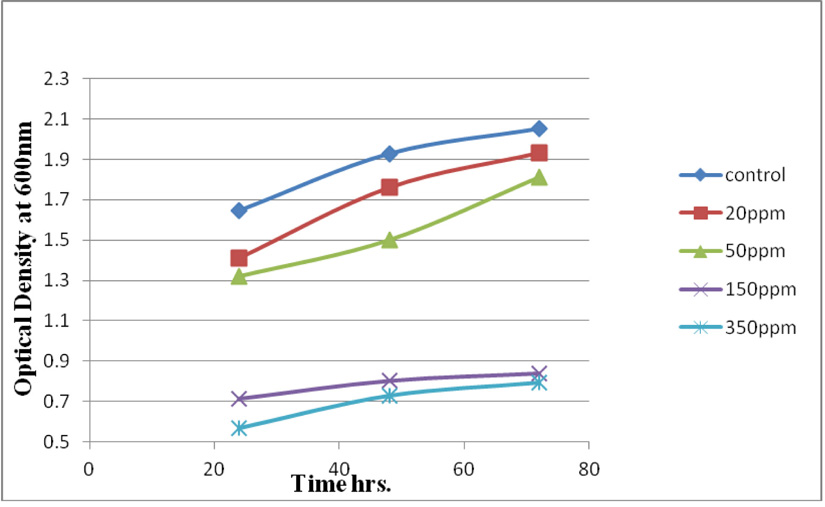
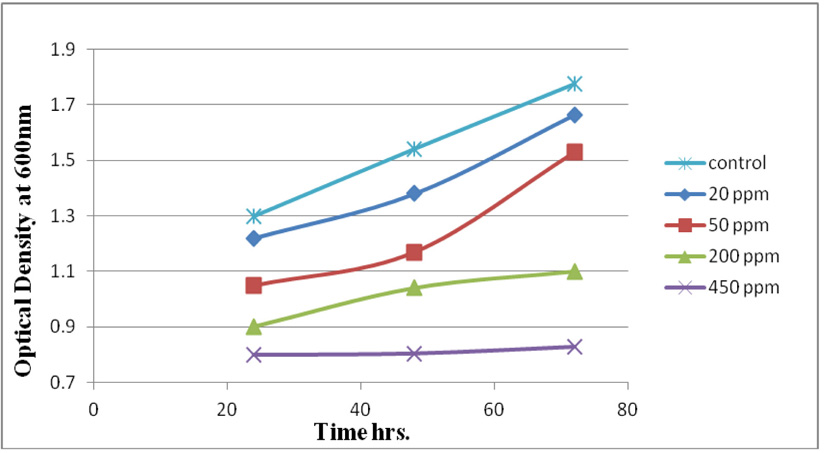
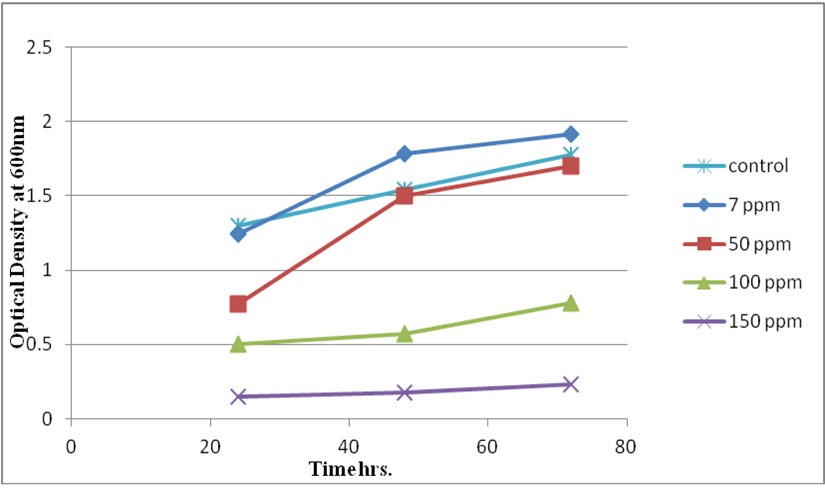
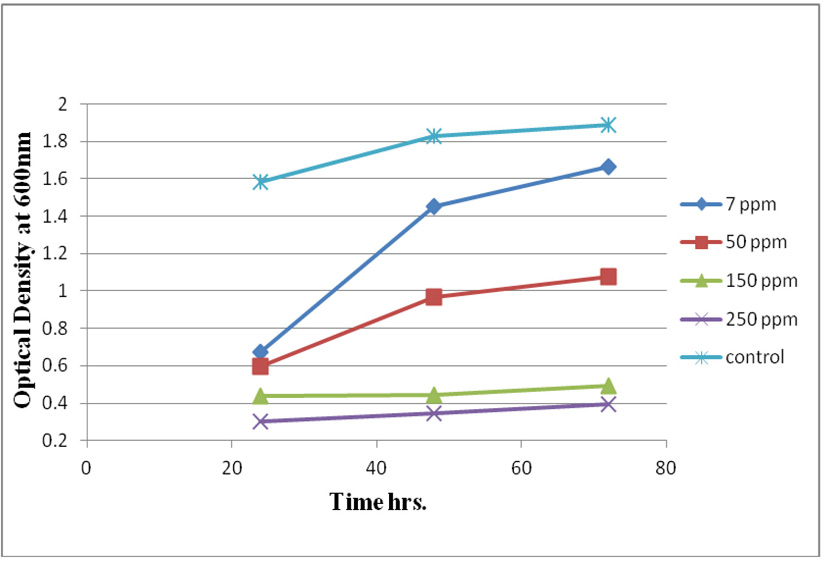
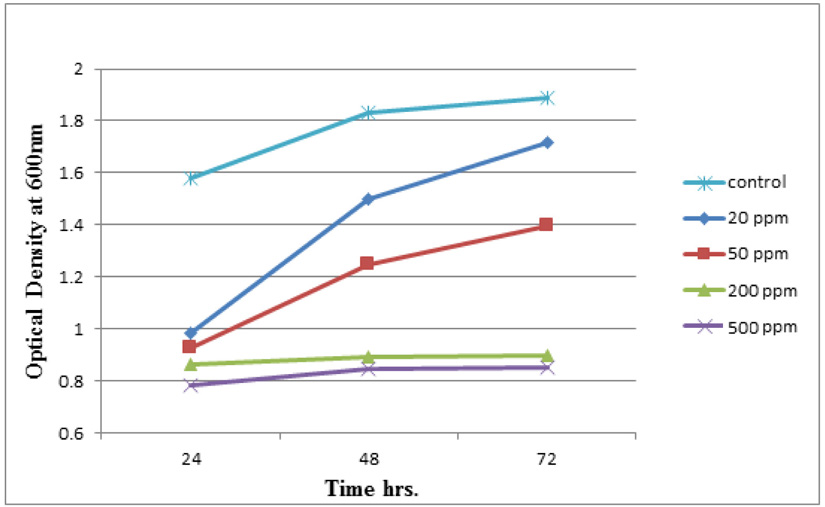
Measuring absorbance at 600 nm using the spectrophotometer, to test tolerance of the bacterial isolates to heavy metals, and then function of biomass. Growth of the isolates on LB with no metal supplementation (control) and with metal supplementation (test). Results showed that growth curves of isolates (A2, PS and ST) in presence of different Pb (NO3)2 and CdCl2 concentrations ranged from (7-550 ppm) are shown in (Fig. 4-11). Results indicated that growth of all isolates decreased with the increase in concentration of Pb (NO3)2 and CdCl2 compared with control.
Morphological and Biochemical Molecular Identification
The morphological and biochemical characteristics of the bacterial (A2, ST and PS) isolates were tested on the nutrient agar. On the basis of the morphological and biochemical study, A2 and ST isolates were identified as Bacillus cereus, and the isolate PS identified as Pseudomonas aeruginosa. The bacterial isolates were positive for (catalase and citrate) while negative for (indole and oxidase) test.
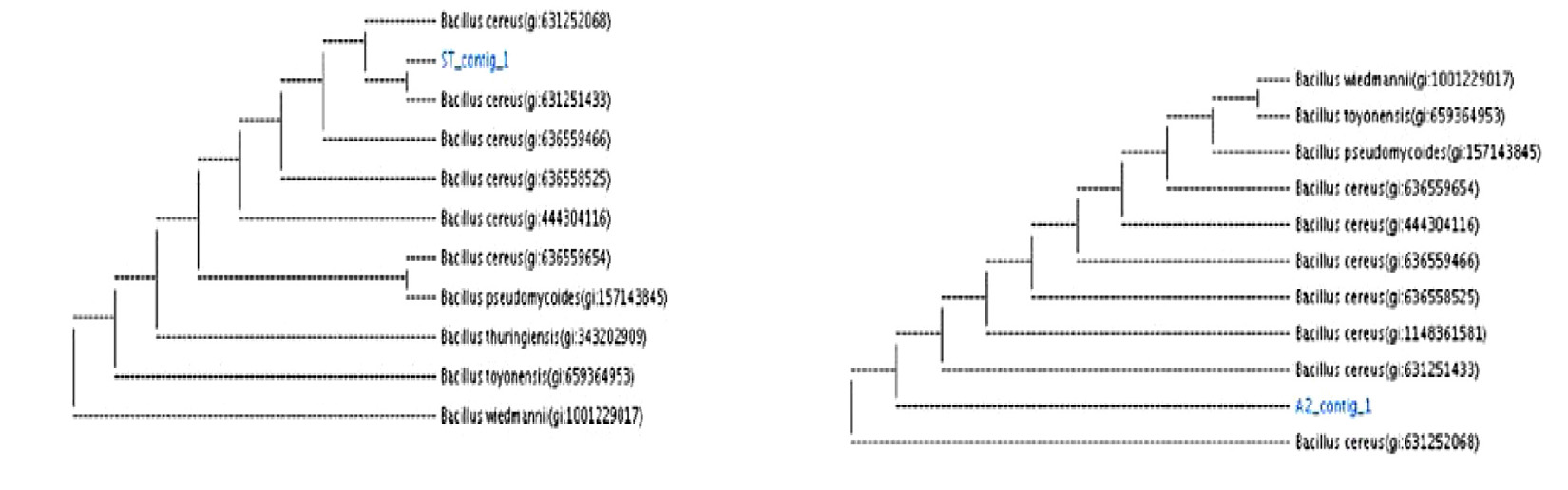
16S rRNA of two strains was sequenced and used to create phylogenetic development tree. Comparative analyses of the sequences from the NCBI databases showed that the strains were closed to the members of the genera Bacillus (Table 4). The highest sequence similarities of the group are ST, B. cereus (99% similarity to B. cereus strain ST; A2, B. cereus (99% similarity to B. cereus strain A2;. Phylogenetic tree of B. cereus ST, A2 shown in (Fig. 12). The sequences were submitted to the NCBI Gene Bank (www.ncbi.nlm. nih.gov) under accession number (MK450303 for B. cereus strain A2) (MK450304 for B. cereus ST).
Table (4):
The closest identified match in the GenBank database.
Accession |
Description |
Query |
Query cover% |
E.Value |
Identity % |
|---|---|---|---|---|---|
MK450303 |
Bacillus cereus |
A2 |
97 |
0.0 |
99 |
MK450304 |
Bacillus cereus |
ST |
98 |
0.0 |
99 |
Plasmid Curing from Three Bacterial Strains
Plasmid curing was carried out to confirm whether-genomic DNA or plasmid encoding of the genes for a resistance of Pb (NO3)2 and CdCl2. Plasmid was cured by the physical agent (elevated temperature 45oC) single colony from B. cereus ST, B. cereus A2 and P. aeruginosa PS strains were picked up and inculcated in NB and incubated at 45 ° C for 24 hrs. One hundred µl from NB speared on NA plate to obtain a single colony. Thirty-two colonies selected and tested on NA medium supplemented with 7ppm concentration of Pb (NO3)2 and CdCl2. Results shown for P. aeruginosa PS and B. cereus A2 strains respectively, that all tested colonies of Pb(NO3)2 and CdCl2 were resistance. Results also, in (Table 5) showed the resistance of cured B. cereus ST for Pb(NO3)2, 32 colonies revealed different degree of resistance 9 colonies lost resistance with (no growth), 14 colonies were weak, 8 colonies had moderate growth. Results in (Fig. 13 and Table 6) showed 9 cured B. cereus ST colonies picked up and tested on nutrient agar medium supplemented with 7ppm CdCl2 revealed that all cured B. cereus ST colonies lost resistance to CdCl2. Nine colonies from cured B. cereus ST were selected for plasmid isolation compared with control.
Table (5):
Growth of B. cereus ST strain in presence of Pb (NO3)2 after plasmid curing for
Mean |
3 |
2 |
1 |
ST strain B. cereus |
|---|---|---|---|---|
+++ |
+++ |
+++ |
+++ |
Control |
– |
– |
– |
– |
1 |
– |
– |
– |
– |
2 |
++ |
++ |
++ |
++ |
3 |
++ |
++ |
++ |
++ |
4 |
++ |
++ |
++ |
++ |
5 |
++ |
++ |
++ |
– |
6 |
++ |
++ |
++ |
++ |
7 |
+++ |
+++ |
+++ |
++ |
8 |
– |
– |
– |
– |
9 |
– |
– |
– |
– |
10 |
– |
– |
– |
– |
11 |
+ |
+ |
+ |
++ |
12 |
+ |
+ |
+ |
+++ |
13 |
++ |
++ |
++ |
++ |
14 |
++ |
++ |
++ |
++ |
15 |
– |
– |
– |
– |
16 |
+ |
+ |
+ |
+ |
17 |
+ |
+ |
+ |
+ |
18 |
+ |
+ |
+ |
+++ |
19 |
+ |
+ |
+ |
+ |
20 |
+ |
+ |
+ |
++ |
21 |
+ |
+ |
+ |
++ |
22 |
++ |
++ |
++ |
++ |
23 |
+ |
+ |
+ |
+ |
24 |
– |
– |
– |
– |
25 |
– |
– |
– |
– |
26 |
– |
– |
– |
– |
27 |
+ |
+ |
+ |
++ |
28 |
+ |
+ |
+ |
++ |
29 |
+ |
+ |
+ |
++ |
30 |
+ |
+ |
+ |
++ |
31 |
++ |
++ |
+++ |
++ |
32 |
Table (6):
Growth of B. cereus ST strain in presence of cadmium CdCl2 after plasmid curing:
3 |
2 |
1 |
ST strain B. cereus |
|---|---|---|---|
+++ |
+++ |
+++ |
Control |
– |
– |
– |
1 |
– |
– |
– |
2 |
– |
– |
– |
3 |
– |
– |
– |
4 |
– |
– |
– |
5 |
– |
– |
– |
9 |
– |
– |
– |
10 |
– |
– |
– |
11 |
– |
– |
– |
16 |
((+++strong, (++) moderate, (+ ) weak and (– )no growth.
Isolation of plasmid from cured B. cereus ST strain
Results in (Table 7) indicated that wild B. cereus ST strain had 2 plasmids (23kb and 564 bp). After plasmid curing all colonies of cured B. cereus ST lost two plasmids while colonies (3, 4, and 5) had one plasmid (23kb).
Table (7):
CdCl2 and Pb (NO3)2 of single colonies after elevated temperature of B. cereus ST.
Plasmid after curing |
Colonies |
|---|---|
1(23kb)
2(564bp) |
Control B. cereus ST |
– |
Cured 1 |
– |
Cured 2 |
1 (23kb) |
Cured 3 |
1(23kb) |
Cured 4 |
1(23kb) |
Cured 5 |
– |
Cured 9 |
– |
Cured 10 |
– |
Cured 11 |
– |
Cured 16 |
Measurement of CdCl2 and Pb (NO3)2 Removal by Bacterial Isolate
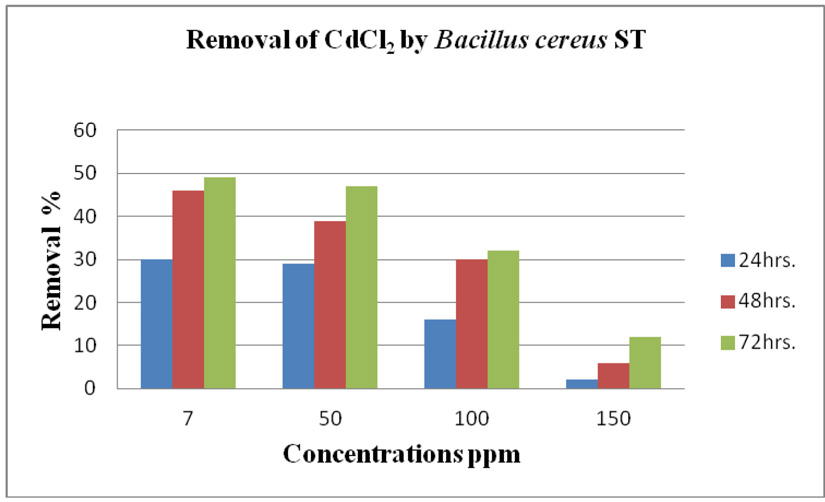
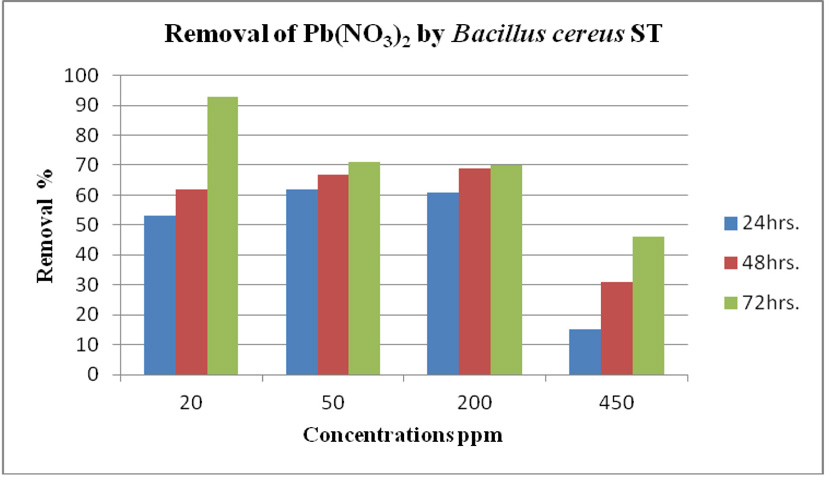
cereus ST, was grown in LB medium supplemented with different concentrations of CdCl2 (7, 50, 100 and 150 ppm) and Pb(NO3)2 (20, 50, 200 and 450 ppm) individually. After time interval 24, 48 and 72 hrs. incubation, cells were separated at 10,000 rpm for 10 min. using centrifuge, then the collected supernatants were measured using ICP-OES which calibrated using un-inoculated medium as a control. Results in (Fig. 14 and Fig. 15) of CdCl2 and Pb(NO3)2 respectively indicated that the highest removal of all concentrations of Pb(NO3)2 and CdCl2 was observed after 72 hrs. and B. cereus ST removal Pb(NO3)2 was estimated (93%, 71%, 70% and 47%) at concentrations (20, 50, 200 and 450ppm) respectively.
While removed CdCl2 at concentration (7, 50, 100 32% and 150 ppm). Removal of 49, 47, 32 and 12%.
Antibiotic resistances
Resistance to antibiotics was determined on Mueller Hinton agar plates. Results in (Table 8) showed the effect of ten antibiotics on the B. cereus indicate that B. cereus A2 was resistance to 5 antibiotic; Metronidazole, Taxo, Tobramycin, Ceftriaxone and Aztreonam while sensitive to Doxycycline with inhibition zone (26 mm). The intermediate inhibition zone for four antibiotics; Tigecycline, Levofloxa-cinycin, Fusidic acid and Vancomycin, (18, 19, 20,17mm). The highest intermediate to Fusidic acid (20 mm). The antibiotic Tobramycin with low inhibition zone (10 mm). B. cereus ST, was highly sensitive to 3 antibiotics; Fusidic acid, Doxycycline and Levofloxacinycin with inhibition zone (23, 24, 25). Intermediate inhibition zone for 3 antibiotics; Tigecycline, Tobramycin and Vancomycin (19, 15, 16 mm) the highest intermediate inhibition zone for Tigecycline (19 mm) and the lowest intermediate inhibition zone for Tobramycin (15 mm) and were no effect for Metronidazole, Taxo, Aztreonam and Ceftriaxone
Table (8):
Inhibition zone of antibiotic susceptibility of the three bacterial strains:
| Bacterial strains | Zone of Inhibition (mm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TGM (15 μg/ml) | LEV (5 μg/ml) | FA (10 μg/ml) | ATM (30 μg/ml) | VA 30 (μg/ml) | Taxo A10 (μg/ml) | TMN (10 mcg) | DOX (30 μg/ml) | MET (50 mcg) | CRO (30 μg/ml) | |
| B. cereus A2 | (I) | (I) | (I) | (R) | (I) | (R) | (R) | (S) | (R) | (R) |
| B. cereus ST | (I) | (S) | (S) | (R) | (I) | (R) | (I) | (S) | (R) | (R) |
| P. aeruginosa
PS |
(R) | (S) | (S) | (R) | (R) | (R) | (R) | (I) | (R) | (I) |
- aeruginosa PS was resistance to 6 antibiotics; Tigecycline, Vancomycin, Taxo A, Tobramycin TMN, Metronidazole and Aztreonam, while sensitive to 2 antibiotic Levofloxacin and Fusidic acid with inhibition zone (20 and 22) respectively. The intermediated inhibition zone for Ceftriaxone and Doxycycline were (16 and 16) respectively
Identify the resistance bacterial strains to heavy metals
Bacterial strains were identified at by the nucleotide blast program at the International Gene-bank site and all strains were identified with 99% compliance: Bacillus cereus ST, Bacillus cereus A2.
cereus is a soil Gram-positive bacterium capable of forming structured multi-cellular communities, or biofilms (Yan et al., 2017). Bacillus cereus can form biofilms on different (surface materials and environmental conditions) (Wijman et al. 2007; Karunakaran and Biggs, 2011). Chien et al., (2013) reported that P. aeruginosa are well known about their biofilm formation are essential factors for resistance heavy metal in Pseudomonas sp. Huang et al., (2013) reported that protection from heavy metal in Bacillus cereus is mostly related to biofilm.
Three bacterial strains (A2, PS and ST) were selected as the most formed biofilms to study of their resistance to the two heavy metals Pb (NO3)2and CdCl2: The resistance of the isolates of CdCl2 and Pb (NO3)2 were studied on nutrient agar plates supplemented primary with 7 ppm concentration. The study showed strong resistance to CdCl2 and Pb (NO3)2 and the ability to grow. The study showed that the maximum tolerance concentrations (MTC) for CdCl2 and Pb (NO3)2 of A2, PS and ST isolates had the ability to grow at different concentrations (7, 10, 20, 50, 100, 120, 150, 200, 250 and 300 ppm) and (7, 20, 50, 100, 150, 200, 250, 300, 350, 400, 450, 500 and 550) for CdCl2 and Pb (NO3)2 respectively. Results showed that the (MTC) of (ST) CdCl2 up to (150 ppm) while the (MTC) was able to grow on Pb (NO3)2 up to (450 ppm). The isolate (A2) showed growth on CdCl2 up to (120 ppm), while the (MTC) was able to grow on Pb (NO3)2 (350 ppm) showed the isolate (PS) able to grow on CdCl2 up to (250 ppm), while the grow on Pb (NO3)2 up to (500 ppm). Rohini and Jayalakshmi, (2015) had published that the MTC value of 100 ppm for cadmium and 500 ppm for lead in Bacillus cereus. Comparatively lower MTC values in the range of 80 ppb to 100 ppm were documented by Tripathy et al., (2011) in Bacillus sp. (Priyalaxmi et al., 2014). Raja et al., (2006) reported that P. aeruginosa (MTC) value for Cd ranged from 100–500 and 100–800 ppm for Pb.
Effect of heavy metals on bacterial growth
Using a spectrophotometer with control samples to study the effect of CdCl2 and Pb (NO3)2 on bacterial growth studied on LB media containing different concentrations (7, 50, 100, 150ppm) and (20, 50, 200, 450ppm) respectively, however the higher the concentration of CdCl2 and Pb (NO3)2 have the less growth due to the toxic effect on bacterial growth. Khatun et al., (2012) studied the effect of two heavy metals, CdCl2 and Pb (NO3)2 on Bacillus cereus growth and showed different growth patterns in the presence of different heavy metals like (Cd and Pb). Rajbanshi, (2008) reported the decrease of the microbial growth with the increase in the concentration of heavy metals indicating a toxicity effect of on the microbial growth. Chien et al., (2013) reported that Pseudomonas aeruginosa are well known about their resistance traits against antibiotics and toxic substances and demonstrated much higher resistance traits against a variety of toxic heavy metals like (Zn, Cd, Ni, Cr, Pb and Cu). However, Huang et al., (2013) reported that protection from Cd2+ in Bacillus cereus is related mainly to biofilm formation. The ability of Bacillus cereus ST to remove heavy metals was tested using the double-thermal plasma induction in the ICP-OES. The results showed that it has a high lead removal capacity of 93% and cadmium to 49%. Ghaima et al., (2013) reported that B. cereus could be used efficiently and effectively in the removal of heavy metals from the polluted environment. Syed and Chinthala, (2015) reported B. cereus exhibited maximum biosorption of lead from heavy metals tested., B. cereus among the isolated strains has shown maximum biosorption of lead (87%), B. subtilis NSPA13 (85%) and B. cereus NSPA8 (78%) respctivly. B. cereus reduced the metal concentration of cadmium by 17%.
16S rRNA
The bacterial isolates were identified biochemically and using 16S rRNA: Some morphological traits, the isolated bacterial colonies were studied. The biochemical tests related to bacterial isolates showed that the two bacterial strains A2 and Bacillus cereus ST were Gram positive while PS Gram negative.
Plasmids curing single colony
To study plasmids curing single colony from B. cereus ST, P. aeruginosa and B. cereus A2 were grown in LB medium at elevated temperature 45°C. One hundred µl were inoculated on a nutrient agar to obtain single colonies. Several colonies (32) were isolated and tested on a nutrient agar plate supplemented with concentration 7ppm on both CdCl2 and Pb (NO3)2 and incubated for 24 hrs. Results showed that 31 out 32 colonies of cured B. cereus ST strain for Pb (NO3)2 that 14 colonies showed weak growth, 8 colonies showed moderate growth, 9 colonies did not grow. Nine colonies were picked up and tested for CdCl2 and showed that all 9 colonies were sensitive (not grow). While cured P. aeruginosa and B. cereus A2 grew naturally when cultured on nutrient agar plate containing 7ppm CdCl2 and Pb (NO3)2. Nine colonies were picked up and studied isolation of plasmid from cured B. cereus ST strain. Results indicated that wild type B. cereus ST strain had 2 plasmids (23kb and 564 bp). After plasmid curing all colonies of cured B. cereus ST lost two plasmids while colonies (3, 4, and 5) had one plasmid (23kb). nald et al., (2003) found that Pseudomonas strains resistant for Cu, Ni, Cd, and Cr, had four plasmids of approximately (20.8, 19.6, 8, and 4.7 kb). They suggested that curing results plasmid DNA conferred nickel and copper resistance, while cadmium and chromium resistance seemed to be encoded by genes on chromosome of bacteria. Al-Charrakh and Al-Enzi, (2016) reported that P. aeruginosa curing showed survived resistance to Pb (NO3). They indicated that the resistance trait was carried on chro-mosome rather than plasmid. Khatun et al., (2012) reveled that B. cereus strain exhibits a single plasmid (48Kb). After plasmid cured B. cereus lost its metal like (Cd2+ and Pb2+) resistance ability. Results indicates that the genes for heavy metals (Cd2+, Cr6+, Ni2+, Co2+) resistance of the isolated strain may be reside on plasmid DNA. However, Alzahrani and Ahamed, (2015) reported that the resistance to three different heavy metals (Ag, Cd and Pb) by Bacillus sp. and showed no plasmid was found resistance trait was carried on chromosome rather than plasmids.
Antibiotics resistances
From antibiotic resistance test of selected bacterial strains ten types of antibiotics were studied
The results showed that the B. cereus ST strain resistant to four antibiotics: Metronidazole (MET), Ceftriaxone (CRO), Taxo (A) and Aztreonam (ATM). While sensitive to three antibiotics, Levofloxacinin (LEV), Fusidic acid (FA), Doxycycline (DOX). B. cereus A2 showed resistance to five antibiotics: Metronidazole (MET), Ceftriaxone (CRO), Taxo (A), Aztreonam (ATM) and Tobramycin (TMN) and sensitive to Doxycycline (DOX). P. aeruginosa PS showed resistance to six antibiotics: Metronidazole (MET), Taxo (A), Aztreonam (ATM), Tobramycin (TMN) and vancomycin (VA), antibiotic sensitive to Levofloxacin (LEV), Fusidic acid (FA), Ceftriaxone (CRO) and Doxycycline (DOX). Ghaima et al., (2013) studied antibiotic resistance of B. cereus and it was resistant to Ceftriaxone. Nakade, (2012) reported that P. aeruginosa was highly sensitive to Levofloxacin. However, Salih et al., (2011) reported that P. aeruginosa resistant to Doxycycline during sensitive Tobramycin. Several isolates were isolated from soil sample located at different locations from Saudi Arabia (Makkah, Taif and Jeddah). Fifty isolates have been tested for formation of biofilm. Results revealed that 3 out of 50 isolates showed high biofilm formation, these three (A2, ST and PS) isolates were tested primary for CdCl2 and Pb (NO3)2 resistances at 7ppm concentration results revealed that isolates were resistance. The maximum tolerance concentration (MTC) of three (A2, ST and PS) isolates studied with different concentrations for CdCl2 and Pb (NO3)2 respectively. Results indicated that MTC values of Pb (NO3)2 were up to (450, 350 and 500 ppm) for ST, A2 and PS isolates respectively. While in CdCl2 the MTC (150, 120 and 250 ppm) for ST, A2 and PS isolates respectively.
hree strains B. cereus A2, B. cereus ST and P. aeruginosa PS that isolated from soil, showed the highest biofilm formation which considered important factor for heavy metals resistance. The biofilm represents a very renewable, promising, cost-effective and easy biotechnology for treatment of wide range contaminated effluents.
The authors thank all members for their help in research, we pleased to provide thanks to King Abdulaziz City for Science and Technology (KACST) for financial support of this research, number of grant (171109000200).
The authors declare no conflict of interest.
- Al-Charrakh, A.H., AlENZI, R.M.J. Heavy metal resistant bacteria pseudomonas aeruginosa as a model. Lap lambert academic publishing, 2013; 978-3-659-36619-2.
- Alloway, B.J. Heavy Metals in Soils 2nd ed. Chapters 6,8,9 and 11. Chapman and Hall, Glasgow, UK, 1995.
- Alzahrani, O.M. and Ahamed, N.T. Isolation and characterization of heavy metal resistant Bacillus subtilis spp. collected from water sources of Taif Province of Saudi Arabia. International Journal of Current Microbiology and Applied Sciences, 2015; 4(6): 350-357.
- Badr, N. B.E., Anwar, A., El-Fiky, A.A., Mostafa, A.R.and Al-Mur, B.A. Metal pollution records in core sediments of some Red Sea coastal areas, Kingdom of Saudi Arabia. Environmental Monitoring and Assessment, 2008; DOI 10.1007/s10661-008-0452-x.
- CHIEN, C.C., LIN, B.C. & WU, C.H. Biofilm formation and heavy metal resistance by an environmental Pseudomonas sp. Biochemical Engineering Journal, 2013; 78: 132-137.
- Christensen, G.D., Simpson, W.A., Younger, J.A., Baddour, L.M., Barrett, F.F., Melton, D.M., Beachey, E.H. Adherence of coagulase negative Staphylococci to plastic tissue cultures: a quantitative model for the adherence of staphylococci to medical devices. Journal of Clinical Microbiology, 1985; 22: 996–1006
- Comte, S., Guibaud, G. and Baudu, M. Relations between extraction protocols for activated sludge extracellular polymeric substances (EPS) and EPS complexation properties Part I. Comparison of the efficiency of eight EPS extraction methods. Enzyme and Microbial Technology, 2006; 38: 237–245
- Decho, A.W. Microbial biofilms in intertidal systems: an overview. Continental Shelf Research, 2000; 20: 1257–1273.
- Gad, G.F.M., El-Feky, M.A., El-Rehewy, M.S., Hassan, M.A., Abolella, H., ElBaky, R.M.A. Detection of icaA, icaD genes and biofilm production by Staphylococcus aureus and Staphylococcus epidermidis isolated from urinary tract catheterized patients. The Journal of Infection in Developing Countries, 2009; 3(5): 342–51.
- Ghaima, K. K., Al Draghi, W. A., Lateef, N. S. Study the heavy metals tolerance, biosorption and antibiotic resistance of Bacillus cereus isolated from diesel fuel polluted soil. International Journal of Biological and Pharmaceutical Research, 2013; 4: 502-506.
- Gomila , M., Gasco´, J., Busquets, A. , Gil, J., Bernabeu,R., Buades, J.M. and Lalucat, J. Identification of culturable bacteria present in haemodialysis water and fluid. FEMS Microbiology Ecology, 2004; 52(2005): 101–114.
- Gutnick, D.L. and Bach, H. Engineering bacterial biopolymers for the biosorption of heavy metals; new products and novel formulations. Applied Microbiology and Biotechnology, 2000; 54:451–460.
- Huang, F., Dang, Z., Guo, C. L., Lu, G. N., Gu, R. R., Liu, H. J., Zhang, H. Biosorption of Cd (II) by live and dead cells of Bacillus cereus RC-1 isolated from cadmium-contaminated soil. Colloids and Surfaces B: Biointerfaces, 2013; 107: 11–18.
- Karunakaran, E. and Biggs, C. A. Mechanisms of Bacillus cereus biofilm formation: An investigation of the physicochemical chara-cteristics of cell surfaces and extracellular proteins. Applied Microbiology and Bio-technology, 2011; 89(4): 1161-1175.
- Khatun, M. A., Bera, P. A., Mitra, D. E., Mandal, A. N. & Samanta, A. M. Estimation of heavy metal tolerance and antibiotic susceptibility of Bacillus cereus isolated from municipal solid waste. International Journal of pharma and bio sciences, 2012; 3: 819-29.
- Kheder AK. Studies on antibiotic resistance by plasmid of Pseudomonas aeruginosa. Ph.D. thesis. College of Education . University of Salahaddin, Iraq, 2002.
- Margeay, M., Nies, D. and Schlegel, H.G. Alcaligenes eutrophus CH34 is a facultative chemolitotroph with plasmid bound resistance to heavy metals. J. Bacteriol, 1985; 162: 328-334.
- McDonald, D.G. and Grandt, A.F. Limestone- Lime Treatment of Acid Mine Drainage-Full Scale. EPA Project Summary (1981). EPA-600/S7-81-033, 1981.
- Morillo , J.A. , Aguilera, M., Ramoz-Cormenzana, A. and Monteoliva Sanchez , M. Production of a metal-binding exopolysaccharide by Paeni-bacillus jamilae using two-phase olive-mill waste as fermentation substrate. Current Microbiology, 2006; 53: 189–193.
- Mulik, A.R and Bhadekar, R.K. HEAVY METAL REMOVAL BY BACTERIAL ISOLATES FROM THE ANTARCTIC OCEANIC REGION. International Journal of Pharma and Bio Sciences, 2017; 8(3): (B) 535 -543, ISSN 0975- 6299.
- Nakade Dhanraj, B. Antibiotic sensitivity of common Bacterial Pathogens against selected Quinolones. Research Journal of Biological Sciences, 2012; 1(1): 77-79.
- Nies, D. H. Microbial heavy-metal resistance. Applied Microbiology and Biotechnology, 1999; l.51: 730–750.
- Outten, F. W., Outten, C. E. and O’Halloran, T. V. Metalloregulatory systems at the interface between bacterial metal homeostasis and resistance.Bacterial stress responses. ASM Press, Washington, DC, 2000; 145-157.
- O’Toole, G.A., Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Molecular Microbiology, 1998; 28(3): 449–61.
- Pandey, G. and Jain, R.K. Bacterial chemotaxis toward environmental pollutants: role in bioremediation. Applied and Environmental Microbiology, 2002; 68: 5789–5795.
- Paul, D., Pandey, G., Pandey, J. and Jain, R. K. Accessing microbial diversity for bioremediation and environmental restoration. TRENDS in Bio-technology, 2005; 23(3): 135-142.
- Post, J. C., Ehrlich, G. D., and Costerton, J. W. BIOFILM REMEDIATION OF METAL CONTAINING WASTEWATER.U.S. Patent No. 8,425,776. Washington, DC: U.S. Patent and Trademark Office, 2013.
- Priyalaxmi, R., Murugan, A., Paul, R. and Raj, K.D. Bioremediation of cadmium by Bacillus safensis (JX126862), a marine bacterium isolated from mangrove sediments. International Journal of Current Microbiology and Applied Sciences, 2014; 3(12): 326-335.
- Raja, E.C. and Selvam, G.S. Plasmid profile and curing analysis of Pseudomonas aeruginosa as metal resistant. International Journal of Environmental Science and Technology, 2009; 6: 259-266.
- Rajeev Kumar, R.M., Tripathi, A.K. Gupta. “Seasonal Variation of heavy metal concen-tration in water of River Yamuna, Allahabad, Uttar Pradesh, India”. International Journal of Current Microbiology and Applied Sciences, 2014; 3(7): 945-949, ISSN: 2319-7706.
- Rohini, R., & Jayalakshmi, S. Bioremediation potential of Bacillus cereus against copper and other heavy metals. International Journal of Advanced Research in Biological Sciences, 2015, ISSN: 2348-8069.
- Salih, H. A., Abdulbary, M., & Abdulride, A. S. Susceptibility of Pseudomonas aeruginosa isolated from urine to some antibiotics. ALQadisiya Journal of Veterinary Medical Science, 2011 10(2): 201-208.
- Sambrook, J., Fritschi, E.F., Maniatis, T. Molecular cloning: a laboratorymanual, Cold Spring Harbor Laboratory Press, New York, 1989.
- Sheng, G.P., Yu, H.Q. and Yue, Z. Factors infl uencing the production of extracellular polymeric substances by Rhodopseudomonas acidophila. International Biodeterioration and Biodegra-dation, 2006; 58: 289–293
- Sutherland, I.W. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology, 2001; 147: 3–9.
- Tay, J.H., Liu, Q.S. and Liu, Y. The role of cellular polysaccharides in the formation and stability of aerobic granules. Letters in Applied Microbiology, 2001; 33: 222–226.
- Thokchom, S., Joshi, S. R. Antibiotic resistance and probiotic properties of dominant lactic microflora from Tungrymbai, an ethnic fermented soybean food of India. The Journal of Microbiology, 2012; 50(3): 535-539.
- Tripathi, M., Vikram, S., Jain, R.K. and Garg, S.K. Isolation and growth characteristics of chromium(VI) and pentachlorophenol tolerant bacterial isolate from treated tannery effluent for its possible use in simultaneous bio-remediation. Indian Journal of Microbiology, 2011; 51(1): 61–69.
- Valls, M. and De Lorenzo, V. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiology Reviews. 2002; 26(4): 327-338.
- Vandamme, P., Pot, B., Gillis, M., De Vos, P., Kersters, K. and Swings, J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiological reviews, 1996; 60(2), 407-438.
- Wijman, J.G.E., de Leeuw, P.P.L.A., Moezelaar, R., Zwietering, M.H., Abee, T. Air-liquid interface biofilms of Bacillus cereus: formation, sporulation, and dispersion. Applied and Environmental Microbiology, 2007; 73:1481–1488.
- Wingender, J., Neu, T.R. and Flemming, H.C. What are bacterial extracellular polymeric substances:. In Microbial extracellular polymeric substances (pp. 1-19). Springer, Berlin, Heidelberg, 1999.
- Yan, F., Yu, Y., Gozzi, K., Chen, Y., Guo, J. H., & Chai, Y. Genome-wide investigation of biofilm formation in Bacillus cereus. Applied and environmental microbiology, 2017; 83(13): 561-17.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.