ISSN: 0973-7510
E-ISSN: 2581-690X
Thermophilic fungi, found in the varied habitats are valuable sources for potential enzymes and other biochemical metabolites throwing challenges to the mycologists. A study of such fungi will not only enables the biochemists, microbiologists, and the biotechnologists to understand the basic molecular process but also search for novel genes, gene products and reactions. In this study, Malbranchea cinnamomea was isolated from goat dung and characterized based on its morphological and physiological characters, and later confirmed molecularly.
Thermophilic coprophilous fungus, Habitats, Temperature, characterization, identification.
Thermophilic fungi grow in a temperature range of 20oC to 60oC1. They are eukaryotic organisms and can grow at high temperatures which makes them easier to study2,3. They are a valuable source for various enzymes with commercial and scientific interest2. Some of the important industrial enzymes isolated from these fungi include L-Asparaginase, cellulases, proteases, amylases, lipases etc. which can be produced on a large scale commercially1. These fungi are also a source of proteins which upon crystallization can help us understand the catalytic mechanism of their intrinsic thermo stability. Another striking feature of these thermostable organisms includes the ability to degrade polysaccharide constituents of biomass1. It is estimated that geothermal habitats are the rich source of thermophiles, especially these areas which are abundant with concentrated sulfur, like acidic soils and acidic thermal waters having a variable pH from neutral to alkaline pH4. In certain other cases these habitats are characterized by presence of certain gases primarily a composition of nitrogen, carbon dioxide, hydrogen sulfide and hydrogen gas4. Life on earth is heterogeneous as is said by Darwin in his theory “survival is the fittest” every flora and fauna adapt to survive in varied conditions found on the earth. These fungi have been reported to be found in natural habitats like soils, air, compost, nesting material, herbivore dung, hot spring, and some of the man- made habitats including pile of wood chips, municipal refugee, peat, pile of hay etc.5 Thermophilic fungi can be grown by enrichment media by providing ambient temperature specific to taxa1. Addition of carbon from different sources shows improved yield6.
The non cellulolytic species utilize xylan, mannose and glucose as the carbon source, in certain other cases a combination of sugars like glucose, sucrose and other sugars in various combinations can also be used pertaining of species6.
Identification always remain a heavy task if only the morphology is used, which enable us to know the genus only. Hence, the ribosomal RNA genes are most commonly used in the identification which acts as an official DNA bar coding marker for species level identification7. A Survey on the natural products secreted by the fungi was conducted by the Journal of Natural Products says that approximately 31% of fungal identification is solely done by morphology, 28% based on the secondary metabolites secreted, 27% by molecular data like ITS and 14% by combining morphological and molecular data like rRNA and protein coding genes8. However accurate identification is required to study further biochemical properties and applications. In the present study thermophilic fungi were isolated from goat dung and characterized.
Chemicals and Reagents
Whatman’s filter paper No. 42, Yeast extract, starch, MgSO4, K2HPO4, sucrose, glucose, Agar-agar and other chemicals obtained from the Himedia labs, Mumbai, India.
Isolation of thermophilic fungi
Media and culture condition
The basal medium Yeast extract starch (YES) agar medium was employed for assessing growth
Starch – 15.0g
Yeast extract – 5.0 g
MgSO4 – 0.5 g
KH2PO4 – 1.0 g
Agar-Agar -20.0 g
Distilled water -1000 ml.
Different source of Herbivore (Cow, Buffalo, Deer, Sheep, Goat, Horse, Elephant etc) dung were collected from Warangal district of Telangana, India in sterilized polyethylene bags and were brought to the laboratory and analyzed for the presence of thermophilic fungi followed by paired plate, serial dilution method as described by Cooney and Emerson 19649 at 45°C temperature. The fungal colonies developed on Petri plates were isolated and transferred to YESA (yeast extract starch agar) slants, allowed to grow and stored at 4°C for further use.
Morphological identification and growth optimization
The isolated fungal colonies were then studied to reveal the Microscopic features of strains such as hyphae, conidiophores, branching conidia, pigmentation etc. were determined using a Olympus make microscope (X40) followed by standard manuals of Cooney and Emerson, 19649 and Mouchacca, 199710. Thermophilic fungi are well recognized group by way of temperature relationship and taxonomic characterization11. Thermophilic fungi grow at elevated temperatures (50°C or above) are often termed as thermophiles2,9.
After incubating the fungus in broth media containing the 50 ml of yeast extract starch without Agar in 100 ml of conical flask for different incubation periods, under the suitable condition at respective temperature, pH and by using different synthetic media included Yeast extract starch (YES), glucose yeast extract (GYE) and czapeck dox modified (CDM). The optimum growth time required for fungus was investigated at various incubation periods (0 day, 3rd day, 6th day, 9th day, 12th day & 15th day). At the end of incubation period the growth of the fungus from day 0 to day 15 was observed by measuring the dry weight of the biomass to understand the growth of the organism, at each interval the culture filtrate was filtered with the help of Whatmann filter paper N0. 42 (measured (w1)) placed on conical funnel . After culture filtration the obtained biomass along with the filter paper was folded and kept for drying in hot air oven (MH Enterprises) and then measured.
Molecular identification
Molecular identification by gene sequencing is important as we can identify the ITS (Internal Transcribed spacer) of fungi that plays a major role in sub generic phylogenetic classification as the locus is specific even within the species12. The total genomic DNA isolated from the mycelial mats using Master pure TM Yeast DNA purification kit as described by Jin et al., 200413. D2 region of large subunit of 28S rDNA gene fragment was amplified as described in earlier work7. Quiagen quick PCR purification kit was used for purification and sequencing. The sequences were then subjected to BLAST16 to identify the nearest taxa and aligned with sequences, belonging to the nearest taxa using CLUSTALW12. The evolutionary history was inferred using the neighbor –joining method. Phylogenetic tree was constructed using the Kimura 2-parameters using MEGA4 software14. Using ITS and rRNA genes to study the taxonomic classification of fungi began 2 decades ago. The fungal DNA sequences for the larger subunit (28S) and the smaller subunits18S) and the ITS (Internal Transcribed Sequences) accompany the fungal identification. ITS is the most useful and required portion of the rRNA cistron8. Protein coding genes are specific to fungal groups and by constructing the phylogenetic trees using the DNA sequences will help in fungal identification15. These genes will give an insight into homology, convergence as it is believed that these genes may be species specific8. Using PCR (Thermo Scientific make) based techniques the detection of intra and inter specific genetic variations can be done that helps in the identification16. These techniques help in evaluating the polymorphisms in the species even with the little differences7. These are the molecular markers that are available for the rapid screening of genetic diversity, these markers allow for high resolution of DNA finger printing17. Neighbour Joining method and MEGA4 software is faster in term of computing the distances14,15,18, estimating genetic distances and diversities, inferring ancestral sequences, computing time trees, and testing selection.
Out of all the samples of various herbivore dung collected, goat dung was selected as a personal choice, through in all the samples thermophilic fungi was observed. An only fungus from goat dung was observed for colony morphology and further molecular identification in the present study.
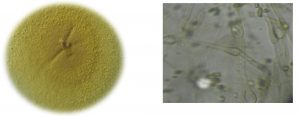
Malbranchea cinnamomea isolated from goat dung on Yeast extract starch media and the colony observed pigmented whitish-yellow. Further identification was done by observing colony features under compound microscope with the help of standard manuals. Colonies on YESA at 45°C were sulfur yellow robust, dense, thick, smooth with a few outward radiating folds, velvetty with coarse, creamy yellow tufts of hyphae. The medium turned black by the diffusion of pigment.
Vegetative hyphae hyaline later became yellowish-brown with racket hyphae, swelling near the septum to a diameter of 9 µm or more. Arthroconidia were observed on curved, lateral branches arising from broader vegetative hyphae, 3-6 µm in diameter (Fig. 1. a & b).
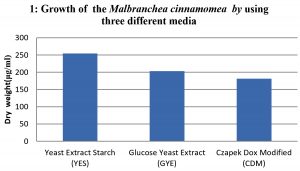
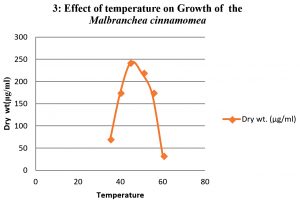
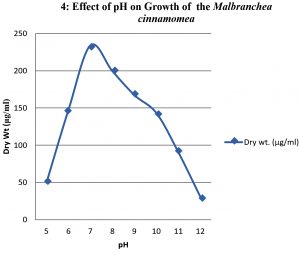
To study the optimum growth conditions of the organism, three different compositions of synthetic Media including Yeast extract starch (YES), glucose yeast extract (GYE) and czapeck Dox modified (CDM) were used and the results are shown in the graph 1. Their period of growth is also studied at different intervals and represented in the graph 2. The factors affecting the growth of the organism were also studied and the results were tabulated specially the effect of pH and the optimum temperature was 45°C (graph 3 & 4). The lowest growth of the fungus was detected at pH 5.0 and temperature 35°C.
When temperature is concerned, the organism shown a growth at temperatures ranging from 35 to 60°C and the growth was ceased above 60°C. In case of pH the growth was optimum at neutral followed by alkaline conditions. The organism was found to be stable upto pH 12.
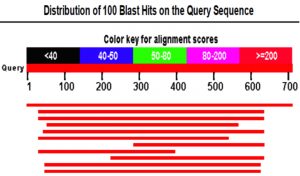
Identification of M .cinnamomea taxonomic status has employed molecular methods involving PCR to resolve genetic variation. A single discrete PCR amplicon band of 711bp was observed when resolved on agarose gel. Consensus sequences of 632bp of D2 region of LSU gene generated sequence gave the detailed data of the sequence obtained primers are presented in chromatograms data (Fig. 2). The obtained DNA sequences data subjected to blast in NCBI and its relatively with ten closest homologs are presented in and alignment view (Fig. 3) the best 10 BLAST hits were taken for phylogentic analysis based on maximum query coverage and maximum identity, one of the sequence from 10 best hit is Malbranchea cinnamomea. Based on nucleotide homology, history of evolutionary distance (table 1) and phylogenetic analysis (Fig. 4) the fungus was confirmed as M. cinnamomea and the accession number given by NCBI Gene Bank data base for Malbranchea cinnamomea isolate GSMBKU as KT279416.1.
Table (1):
Distance matrix method.
GSMBKU_KT279416.1 |
0.05 |
0.05 |
0.05 |
0.05 |
0.05 |
0.05 |
0.05 |
0.05 |
0.05 |
0.05 |
|
KJ834332.1 |
0.68 |
0.00 |
0.05 |
0.06 |
0.00 |
0.05 |
0.05 |
0.05 |
0.05 |
0.05 |
|
KJ834368.1 |
0.68 |
0.00 |
0.05 |
0.06 |
0.00 |
0.05 |
0.05 |
0.05 |
0.05 |
0.05 |
|
JF412018.2 |
0.68 |
0.68 |
0.68 |
0.05 |
0.05 |
0.05 |
0.06 |
0.05 |
0.05 |
0.05 |
|
KJ563258.1 |
0.74 |
0.69 |
0.69 |
0.69 |
0.06 |
0.05 |
0.05 |
0.06 |
0.05 |
0.05 |
|
KJ834339.1 |
0.68 |
0.00 |
0.00 |
0.68 |
0.69 |
0.05 |
0.05 |
0.05 |
0.05 |
0.05 |
|
JF922020.1 |
0.76 |
0.74 |
0.74 |
0.74 |
0.73 |
0.74 |
0.05 |
0.05 |
0.05 |
0.06 |
|
EF550978.1 |
0.69 |
0.75 |
0.75 |
0.56 |
0.75 |
0.75 |
0.78 |
0.05 |
0.05 |
0.05 |
|
KP828175.1 |
0.68 |
0.78 |
0.78 |
0.75 |
0.63 |
0.78 |
0.74 |
0.70 |
0.06 |
0.05 |
|
KP278210.1 |
0.78 |
0.80 |
0.80 |
0.76 |
0.79 |
0.80 |
0.70 |
0.69 |
0.71 |
0.05 |
|
KP278209.1 |
0.70 |
0.75 |
0.75 |
0.70 |
0.69 |
0.75 |
0.74 |
0.71 |
0.69 |
0.68 |
Accession |
Description |
Max score |
Total Score |
Query Coverage |
E Value |
Max. Identity |
|---|---|---|---|---|---|---|
KT279416.1 |
Malbrancheacinnamomea isolate GSMBKU5.8S ribosomal RNA |
1314 |
1314 |
100% |
0. 0 |
100% |
KJ834332.1 |
Uncultured fungus clone Z1E-1 18S ribosomal RNA |
1018 |
1018 |
84% |
0. 0 |
97% |
KJ834368.1 |
Uncultured fungus clone Z1C-52 18S ribosomal RNA |
1005 |
1005 |
84% |
0. 0 |
97% |
JF412018.2 |
Malbrancheacinnamomea strain CBS 343.55 18S ribosomal RNA |
876 |
876 |
72% |
0. 0 |
98% |
KJ563258.1 |
Malbrancheacinnamomea isolate CM-10T 5.8S ribosomal RNA |
874 |
874 |
82% |
0. 0 |
94% |
KJ834339.1 |
Uncultured fungus clone Z1E-9 18S ribosomal RNA |
846 |
846 |
71% |
0. 0 |
97% |
JF922020.1 |
Malbrancheacinnamomea strain CBS960.72 5.8S ribosomal RNA |
616 |
616 |
49% |
3.00E-172 |
99% |
EF550978.1 |
Malbrancheaflava isolate MTCC 4887; 5.8S ribosomal RNA gene |
558 |
558 |
51% |
5.00E-155 |
94% |
KP828175.1 |
Uncultured fungus clone CII-K39 5.8S ribosomal RNA |
466 |
466 |
57% |
3.00E-172 |
88% |
KP278210.1 |
Gymnoascus sp. NZ-2015b strain S-5c 18S ribosomal RNA |
466 |
466 |
81% |
3.00E-172 |
82% |
KP278209.1 |
Gymnoascus sp. NZ-2015b strain S-4c 18S ribosomal RNA |
466 |
466 |
81% |
3.00E-172 |
82% |
ACKNOWLEDGMENTS
Shanthipriya Ajmera acknowledges U.G.C (Rajiv Gandhi National Fellowship) for the financial assistance. Authors are thankful to the Head, Department of Microbiology, Kakatiya University for providing the necessary facilities.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SA and VR were involved in collection of the samples and molecular analysis of the fungal species. VKR and RM have done the bioinformatic analysis of the data obtained from sequencing. SG was involved in conceptualization of idea and wrote the manuscript.
FUNDING
The funding was obtained from the University Grants Commission, New Delhi, India UGC RGNF-JRF/SRF (F1-17.1/2012-13/RGNF-2012-13-ST-AND-28098).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
- Rajkumar Salar-CRC Press section 1, Chapter 1-4, A text Book on -Thermophilic Fungi: Basic Concepts and Biotechnological Applications, 2018; 10-144.
- Ramesh Maheshwari, Girish Bharadwaj, Mahalingeshwara K. Bhat, Thermophilic Fungi: Their Physiology and Enzymes -Microbiology and Molecular Biology, Reviews-American Society for Microbiology.
- Saroj Ahirwar, HemantSoni, BhanuPratap Fungi in Unusual Environments Isolation and screening of thermophilic and thermotolerant fungi for the production of hemicullases from heated environments, Mycology An international Journal on Fungal Biology, 2017; 8: 125-134.
Crossref - G. C. Ainsworth, Alfred S. Sussman, A Text book on The Fungal population: An Advanced Treatise- Volume III the Fungal Population, Academic press,New York, London, pg 105 – 125
- Preeti Singh Parihar and VibhutiRai . Isolation, screening and selection of xylanase producing thermophilic fungi from Raipur Research in Biotechnology, 2015; 6(4): 27-37, -791X .
- Gregory M. Mueller, Gerald F.Bills Mercedes, S.Foster. A text book on Biodiversity of Fungi: Inventory and Monitoring Methods, 2004; 59-77, 467-500
- Matsuda Y, Hijii N; Characterization and identification of Strobilomyces confusus ectomycorrhizas on Momi fir by RFLP analysis of the PCR-amplified ITS region of the rDNA; J. For Res.; 1999; 4: 145- 150.
Crossref - Huzefa A. Raja, Andrew N. Miller Cedric J. Pearce, Nicholas H. Oberlies. Fungal Identification Using Molecular Tools: A Primer for the Natural Products Research Community. J. Nat. Prod., 2017; 80(3): 756–770.
Crossref - Cooney, D.C., and R. Emerson, W.H. Freeman and Co., San Francisco In: Thermophilic fungus: An account of their Biology, Activities and Classification, 1964; 1-188.
- Mouchacca J, Thermophilic Fungi: Biodiversity and Taxonomic Status. Cryptogamie, Mycologi, 1997; 18: 19-69.
- Jarvis BW, Wickes B L, Hoffman LM;direct genomic DNA sequencing for rapid fungal identification (ASM meeting Orlando); J. Clin. Microbiol., 2010; 48(3): 741-752.
- Sarkar S, Girisham S, Reddy S M;Identification of three fruit rot fungi of banana by 28S ribosomal DNA sequencing; IJB, 2013; 2: 422-429.
- Jin. J. J. Clin. Microbiol., 2004; 42: 429.
- Sudhir Kumar, Glen Stecher, Michael Li, Christina Knyaz, Koichiro Tamura MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution, 2018; 35: 1547–1549.
Crossref - Morgenstern. I., Justin Powlowski,Nadeeza Ishmael, Corinne Darmond, Sandrine Marqueteau., A molecular phylogeny of thermophilic fungi. Fungal Biology, 2012; 116(4): 489-502.
Crossref - Paromik Bhattacharyya, Subhabrata Ghosh, Swati Sen Mandi, Suman Kumaria, Pramod Tandon. Genetic variability and association of AFLP markers with some important biochemical traits in Dendrobiumthyrsiflorum,a threatened medicinal orchid. South African Journal of Botany, 2017; 109: 214-222.
Crossref - Ulrich G. Mueller, L. La Reesa Wolfenbar AFLP genotyping and fingerprinting. Trends in Ecology and Evolution, 1999; 10: 389-394.
Crossref - Irina, S. Druzhinina, G.Alexi Kopchinskiy, Monika Komon, John Bissett, George Szakacs and P. Christian Kubicek, An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genetics and Biology, 2005; 42: 813-828.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.