ISSN: 0973-7510
E-ISSN: 2581-690X
Currently, there is an increase prevalence of antibiotic-resistant bacteria worldwide. Therefore, the need for characterization of naturally occuring antibiotics with less antibiotic resistance is required. Soil resources contains valuable antibiotic producing microorganisms that increasingly being utilized for the production of suitable antibiotics. Therefore, this study aimed at identifying an antibiotic bacteria with ability of producing antibiotic that is isolated from soil samples collected from Al Zarqa provenance, an arid area in Jordan. Morphological and biochemical characterization of the isolates were carried out and found that all of the isolates belong to Bacillus genus. Further confirmation of the characterization of the bacteria was done by ribosomal RNA and PCR. The results reveal that the isolates represent Basilluslicheniformis. These bacilli were further investigated for antimicrobial activities against 6 ATCC human pathogens viz., S. aureus, S. pneumonia, Salmonella typhi., E. coli, P. mirabels and E. cloacae. Additionally, the results of Gas Chromatography Mass Spectrometry (GCMS) of ethyl acetate extracts for B. licheniformis secondary metabolites showed that they contain two main antimicrobial compounds namely Pyrrolo [1, 2-a] pyrazine-1, 4-dione,hexahydro and Trans-13-octadecenoic acid. The present work maybe suggests that soil isolates from the studied arid area include antibiotic producing strains that can be utilized commercially.
Bacillus, Antibacterial activity, 16S rRNA, GCMS
The field of medicine and pharmacology are currently revolutionized thanks to the invention of antibiotics that successfully that has been fruitful in combating many infectious diseases. However, many infectious microbes have become increasingly resistant to most available antibiotic due to mainly the extensive these antimicrobial medicine1. Varied genetic mechanisms, such as mutation, genetic transfer, and epigenetic, have been adopted by resistant pathogens to make them successful adapted to antibiotics2. Antibiotics and antimicrobial metabolites extracted from microorganisms that inhabit environmental habitats, such soil, are providing an important role in fighting microbial disease and shown to be a promising source of new novel antibiotics3. Natural soil in general has a high biodiversity of bacteria and provide an important resource for these microbial diversity for possible novel antibiotics4. In fact, several medicinally new antibiotics have been characterized and identified from bacterial that inhabit varied natural soil, isolated from different soil samples5,6,7. The Bacillus is a heterogenous genera of bacteria with species that contain enormous antimicrobial compounds that act as agent of fighting several microbial diseases8. Among the most important species of bacteria that produce medically importan antibiotics are beloging to the genus Bacillus9. These species and other of Bacillus are among the most abundant bacterial strains found in soil with the ability of endospore-forming and Gram positive bacteria. Many investigations have been carried out to isolate different strains of terrestrial bacillus and identify their antimicrobial activities. This study attempts using biochemical and 16S rRNA to identify the microbial isolates from an arid are, Al Zarqa, of Jordan and to determine their antimicrobial activities.
Microorganisms
Based on the study of Massadeh and Mahmoud10, new isolates of antagonistic strains were gathered from soil that was collected from the Hashemite University Campus area and identified as Bacillus species. Three isolates were chosen for further screening of antimicrobial activities and possible production of secondary metabolites. Each isolate was maintained on nutrient agar slants, and preserved at 4°C.
Isolates identification
The isolates were characterized to the genus level by based on their morphology and biochemical characteristics11. The culture characteristics include colony morphology (size, opacity, form, elevation and margins), Gram reaction, spore formation, motility test, and Biochemical reactions. Further identification to the species level was achieved by using Microgen Bacillus-IDkits (Microgen bioproducts, UK) provided with Microgen identification software (MID-60).
Molecular identification of the isolates
DNA extraction
The DNA of each bacterial strains was isolatedfrom a 24 hours old pure culture based on methodology of bacterial DNA extraction kit (Wizard®, Promega, USA). The DNA was kept frozen until sequencing.
16S rRNA gene amplification and sequencing
Two universal PCR primers were manufactured to amplify approximately 1,300 bp a region of 16S ribosomal RNA gene: the forward primer 63f (5`-CAG GCC TAA CAC ATG CAA GTC-3`) and the reverse primer 1387r (5`-GGG CGG WGT GTA CAA GGC-3`).The PCR mixtures contained approximately 2 μl of DNA and 1 μl of each primer and 12.5 μl of DNA master mix (Promega, USA) and about 8 μl of double distelled water. The reactions of PCRwere performed with a cycler (biorad thermal cycler). The cycler was programmed to perform 94°C for 30 min, followed by 25 cycles consisting of 94°C for 45 s, 55°C for 45 s, and 72°C for 90 s. followed by a final extension step of 10 min. at 72°C. PCR products were visualized by 1% agarose gel electrophoresis. The PCR product was properly labeled and sent to Princess Haya Biotechnology Center (Irbid-Jordan) for being identified by sequencing of the PCR products.
Antibacterial activity
Cell-free extract preparation
Each isolate was cultivated in a flask containing a 100ml of nutrient broth. The flasks were incubated at 30°C in a shaking incubator running at 150 rpm for 3 days and then subjected to centrifugation and then filltered with standard memebrane.
Ethyl acetate extract preparation
To concentrate the isolates metabolites, each extract was mixed with ethyl acetate at a ratio of 4:1. The mixture was agitated at 100 rpm for 2 hrs in an incubator shaker (Human Lab, Korea). The organic phase was concentrated by evaporating the ethyl acetate at 45°Cand the resulting extract was stored at 4°C until use10.
Antimicrobial test
The inhibitory effect of each bacterial extract was tested against different pathogens using agar-well diffusion method. The tested pathogenic microorganisms include 2 Gram-positive bacteria (Streptococcuspneumonia ATCC 6303 and Staphylococcus aureus ATCC 11632) and 4 Gram-negative bacteria (Proteus mirabilis ATCC, Enterobactercloacae ATCC 13182., Escherichia coli ATCC 10145, and Salmonella typhi. ATCC 13076). A suspension of each test pathogens (0.1 ml) of an O.D600 of 0.4 was spread on NA plates, and wells of 6mm. A 40 μl of extract was placed into the wells, and the plates were placed under 37°C for about 24 hours. The diameter of affected zone was estimated with calipers as mm. The antibiotic Ciprofloxacin (5 mcg) was used as a control.
Characterization of antibacterial metabolites
Proteolytic digestion
For testing proteolysis of metabolites was employed with a slight modification accordingto the procedure describe eleswhere12. One milliliter of eythl acetate extract showing antibacterial activity was subjected to proteolytic digestion using a 100 mg/ml proteinase K. The samples then were incubated overnight at 56°C. After incubation, samples were tested for antibacterial activity by the agar well diffusion method and a sample of ethyl acetate extract without proteinase K was used as a negative control.
Thermostability test
Each extract was incubated at 80°C for 1 hr. and the tested for antibacterial activity by the agar welldiffusion method. A sample of ethyl acetate extract without heat treatment was used as a negative control.
Detection of antibacterial compounds by Gas chromatography/Mass spectrometry (GC-MS)
Antibacterial metabolites in ethyl acetate extract were characterising using GC-MS by inserting a 1 μl solution into an Agilent 6890 GC detector GC-MS. The temperature level procedure was set at 80°C for about 3 min and then was increased to 80 to 500°C. Chemistation system was used to compare the percntage of mass to charge using standared spectrum library13.
Morphological characteristics and biochemical activities
The major aim of current research was to identify and characterize bacterial strains isolated from soil Al Zarqa arid area that has the ability of producing natural antibiotic. The soil in this area was chosen as it contains large bacterial community with potential of antibiotic production10. The results revealed 3 isolates with mainly yellow-white opaque morphological characteristic for studied colonies in accordance with similar studies that suggested soil as an important natural resource of medically important bacteria14. In agreement with similar the resultsthe biochemical tests of the four isolates revealed that all isolated were positive for catalase production15. Additionally, Microgen Bacillus-ID identified 3 possible isolates with varied percentage of probability: B. subtilis, 82.7%, B. licheniformis with both 37.9% and 80.2% (Table 1). Moreover, the results showed that are positive to citrate utilization and have significant capability of fermentation of the sugars cellobiose, mannitole, mannose, salicin, sucrose, trehalose and xylose (Tables 2).
Table (1):
Identification of species of the four isolates by Microgen Identification System Software (MID-60).
Isolate |
Identification |
Percent probability |
Identification comments |
|---|---|---|---|
1 |
B. subtilis |
82.69% |
Acceptable Identification, additional tests may improve the identification |
2 |
B. licheniformis |
37.94% |
Identification to the species level will require additional tests |
3 |
B. licheniformis |
80.18% |
Acceptable Identification |
Table (2):
Biochemical reactions of using the test of Microgen Biochemical ID Software Microgen GN-A(after 24 h).
Reaction name |
1 |
2 |
3 |
|---|---|---|---|
ARA |
+ |
+ |
+ |
CEL |
+ |
+ |
+ |
INO |
– |
+ |
– |
MAN |
+ |
+ |
+ |
MNS |
+ |
+ |
+ |
RAF |
– |
– |
– |
RHF |
– |
– |
– |
SAL |
+ |
+ |
+ |
BOR |
+ |
+ |
+ |
SUC |
+ |
+ |
+ |
TRE |
+ |
+ |
+ |
XYL |
+ |
+ |
+ |
ADO |
– |
– |
+ |
GAL |
+ |
– |
+ |
MDM |
– |
– |
– |
MDG |
+ |
+ |
+ |
INV |
– |
– |
+ |
MLZ |
– |
– |
– |
INO |
—- |
—- |
—- |
ONPG |
+ |
+ |
+ |
ARG |
– |
– |
– |
CIT |
+ |
+ |
+ |
VP |
—- |
—- |
—- |
NIT |
—- |
—- |
—- |
Genetic characterization of bacterial strains by using ribosomal RNA
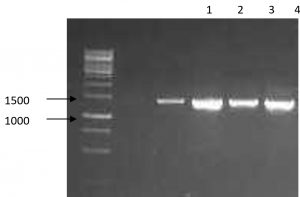
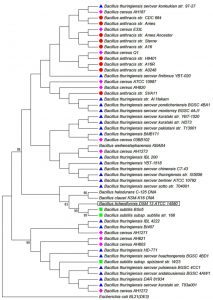
To confirm the identification and the classification of the studied strains all isolates were subjected to advanced identification using 16S rRNA sequencing16. DNA of each isolate was extracted, amplified in PCR, checked in electrophoresis and finally sequenced. Fig. 1 and Table 3 shows the main products of gel electrophoresis and identification according to 16S RNA sequencing. Moreover, a phylogenetic tree, constructed by Neighbor-joining, showed a four major group of Bacillus strains that clustered based on their sequence similarity of the 16S rRNA gene from all strains17 (Fig. 2). This clustering method is preferable to the conventional biochemical tests as it reveals the identification of isolates within 2-3 days compared to several weeks18. In agreement with several previous reports our results revealed that ribosomal RNA analysis is an efficient method for classification of bacteria and provide support for earlier studies which have shown that Bacillus species as common bacteria occur in soil19.
Table (3):
Classification of bacterial strains according to 16S rRNA (results obtained from BLAST/NCBI).
Isolate |
Classification |
Ident |
Accession No. on BLAST |
|---|---|---|---|
1 |
B. licheniformis DSM 13 |
87% |
NR 118996.1 |
2 |
B. licheniformis DSM 13 |
90% |
NR 118996.1 |
3 |
B. licheniformis DSM 13 |
97% |
NR 118996.1 |
Fig. 2. Phylogenetic tree constructed by Neighbor-joining based on the sequences of the 16S rRNA gene from 50 Bacillus strains (Wang and Ash, 2015)
Antimicrobial activity
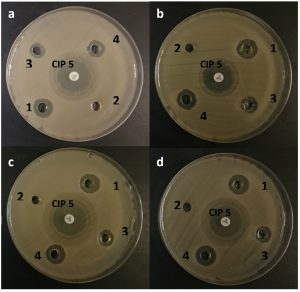
The antimicrobial activity of the 3 isolates was studied against tested human pathogens. The results revealed that the ethyl acetate extract of the 3 isolates of B. licheniformis DSM 13 have an inhibitory effect against all test pathogens except for E. coli ATCC 10145 and Salmonella typhi. ATCC 13076 (Table 4; Fig. 3). It was found that the highest antimicrobial activitywas achieved by the extract B. licheniformis DSM 13 (isolate 1) against S. Aureus ATCC 11632 where the inhibition zone was 13.33 ± 0.58 mm, while the lowest antimicrobial activity was exhibited by B. licheniformis DSM 13 (isolate 2) extract against E. cloacae ATCC 13182, P. mirabilis ATCC 12453 and S. pneumonia ATCC 6303 with inhibition zones of 10.33 ± 0.58 mm (Table 4). Previous reportesindicated that B. licheniformis is widely distributed in nature and can readily be isolated from soils3. Moreover, B. licheniformis has been applied widely for producing extracellular enzymes, antibiotics, and special chemicals with a low risk of adverse effects to human health or the environment20.
Table (4):
Antimicrobial activity of the three isolates of B. lichinifomis against test pathogens.
| Pathogen | Diameters of inhibition zone (mm) by the four isolates | CIP 5 | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| E. cloacae ATCC 13182 |
13.00 ± 1.00 | 10.33 ± 0.58 | 12.33 ± 0. 58 | 21.33 ± 0.58 |
| P. mirabilis ATCC 12453 |
12.67 ± 0.58 | 10.33 ± 0.58 | 11.67 ± 0.58 | 22.00 ± 1.00 |
| E. coli ATCC 10145 |
0 | 0 | 0 | 0 |
| Salmonella typhi ATCC 13076 |
0 | 0 | 0 | 0 |
| S. pneumonia ATCC 6303 |
12.67 ± 0.58 | 10.33 ± 0.58 | 11.67 ± 0.58 | 20.33 ± 0.58 |
| S. aureus ATCC 11632 |
13.33 ± 0.58 | 11.67 ± 0.58 | 12.67 ± 0.58 | 22.00 ± 1.00 |
Fig. 3. Antimicrobial activity of the 3 isolates against the test pathogens: In this figure: a: Enterobacter sp., b: P. mirables, c: Staphylococcus sp., d: Streptococcus sp. No. 2 well and CIP 5 (Ciprofloxacin 5 μg/disc) were used as a negative and positive control.
Characterization of ethyl acetate extract of B. licheniformis isolates
The extracts were subjected to heat treatment and proteolysis by enzymes. The results revealed that heat treatment of the etheyl acetate extracts of the 3 isolates did not reduce the antibacterial activity. The activity could still be demonstrated even after subjecting the extracts to 80°C for 1 hr. Some reportes indicated that the thermal stability of many antibacterial compounds isolated from B. licheniformis21,22.Other, hovere, claimed that the most metabolites of Bacillus sp. were proved to be stable at different temperatures23. On the other hand, incubating the extracts supplemented with Proteinase K did not affect the antibacterial activity of the extracts of the 3 active isolates suggests that the active compounds are not proteinaceous in nature.
Table (5):
Gas chromatogram results of the major compounds of the three active isolates.
isolate |
Compound |
Retention time (min) |
|---|---|---|
1 |
Pyrrolo [1, 2-a] pyrazine-1, 4-dione, hexahydro Trans-13-octadecenoic acid |
12.397 16.127 |
2 |
Pyrrolo [1, 2-a] pyrazine-1, 4-dione, hexahydro Trans-13-octadecenoic acid |
12.397 16.127 |
3 |
Pyrrolo [1, 2-a] pyrazine-1, 4-dione, hexahydro |
12.397 |
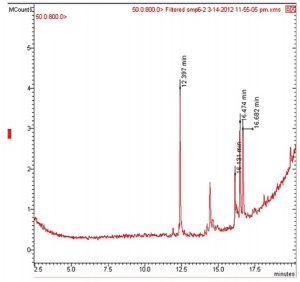
GCMS analysis
Chemical investigations for the 3 ethyl acetate extracts based on GCMS indicated the identification of 2 major compounds (Table 5): (1) Pyrrolo [1, 2-a] pyrazine-1, 4-dione, hexahydro with a retention time of 12.397 min. It was evident that this cyclic peptide is produced by some Streptomyces strains and sponge-associated marine bacteria, in addition to some endophytes. Some researchers reported the isolation of Streptomyces strain MUSC149T mangrove soil with a strong antioxidant activity24. The chemical analysis this strain’s extract revealed the identification of Pyrrolo[1,2-a] pyrazine-1,4-dione, hexahydro as the antioxidant agent. On the other hand, some researchersidentified the presence of this compound in the extract of an Antarctic endophytic fungus exhibiting a strong antibacterial activity against E. coli, Pseudomonas aeruginosa and Enterococcus faecalis13. Moreover, this compound was produced by sponge associated bacteria (SAB) that was capable of inhibiting Vibrio alginolyticus25. (2) Trans-13-octadecenoic acid with a retention time of 16.127 min. this Trans fatty acid was detected in tuber extracts of Solena amplexicaulis plant and has been reported as anti-inflammatory and cancer compound26.However, other scientists identified this compound as an anti-inflammatory and antimicrobial compound22.
The miss use of antibiotics has led to the evolution of pathogens with resistance to major available antibiotics. Accordingly, natural habitat such as soil are emerging procedure for the synthesis of new, safe and developed antibiotics. Three antagonistic bacterial strains were isolated from the soil of an arid area of Hashemite University and identified to be B. licheniformis based on biochemical tests and 16S rRNA gene sequencing with antibacterial activity against important human pathogens. Our finding promote that the bacterial strains obtained from the studied soil sables can be exploit commercially. Based on our study, we recommend further research that aim to use MIC to determine antibacterial activities, in addition to molecular docking and in vivo investigations to identify active compounds.
ACKNOWLEDGMENTS
We acknowledge the Hashemite University of Jordan for facilitating the conduct of this article.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All Authors contributed equally to literature collection, designing and writing the manuscript.
FUNDING
This work was financially funded by the Deanship of Research of the Hashemite University. Grant number is 1600204/9.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Cars O, Hogberg LD, Murray M, et al.Meeting the challenge of antibiotic resistance. BMJ. 2008;18:337:a1438.
Crossref - Bachoual R, Tankovic J, and Soussy C.J. Analysis of the mutations involved influoroquinolone resistance of in vivo and in vitro mutants of Escherichia coli. Microb Drug Resist. 1998;4:271-276.
Crossref - Kumar D, Kumar S. Antimicrobial metabolites and antibiotics obtained from different environmental sources. Int J Pharm Res Allied Sci. 2016;5:85-90.
- Amin M, Rakhisi R, Ahmady AZ. Isolation and Identification of Bacillus species from soil and evaluation of their antibacterial properties. Avicenna J Clin Microb Infec. 2015;2:e23233.
- Griffith RS. Introduction to vancomycin. Rev Infect Dis. 1981;3:200-204.
Crossref - Umezawa H, Ueda M, Maeda K, et al. Production and isolation of a new antibiotic: kanamycin. J Antibiot. 1957;10:181-188.
- Staunton J, Wilkinson B. Biosynthesis of erythromycin and rapamycin. Chem Rev. 1997;97:2611-2630.
Crossref - Sumi CD, Yang BW, Yeo I-C, Hahm YT. Antimicrobial peptides of the genus Bacillus: a new era for antibiotics. Can J Microbiol. 2015;61:93-103.
Crossref - Yilmaz M, Soran H, Beyatli Y. Antimicrobial activities of some Bacillus spp. Strain isolated from the soil. Microbiological Research. 2005;161:127-131.
Crossref - Massadeh MI, Mahmoud S. Antibacterial activities of soil bacteria isolated from Hashemite University area in Jordan. Jordan J Biol Sci. 2019;12:503-511.
- Brawn A. Benson’s Microbiological Applications: Laboratory Manual in General Microbiology, 9th edition, McGraw-Hill, New York. 2004.
- Boottanun P, Potisap C, Hurdle JG, Sermswan RW. Secondary metabolites from Bacillus amyloliquefaciens isolated from soil can kill Burkholderia pseudomallei. AMB Express. 2017;7:16-27.
Crossref - Melo I, Santos S, Rosa L, et al. Isolation and biological activities of an endophytic Mortierella alpine strain from the Antarctic moss Schistidium antarctic. Extremophiles. 2014;18:15-23.
Crossref - Kaur S, Kaur J, Pankaj PP. Isolation and characterization of antibiotic producing microorganisms from soil samples of certain area of Punjab region of India. Int J Pharm Clin Res. 2014;6:312-5.
- Yunus FN, Khalid ZZ, Rashid F, Ashraf A, Iqbal MN, Hussain F. Isolation and screening of antibiotic producing bacteria from soil in Lahore city. PSM Microbiology. 2016;1:1-4.
- Janda JM, Abbott SL, 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45(9):2761-2764.
Crossref - Wang A, Gavin J. Ash. Whole genome phylogeny of Bacillus by feature frequency profiles (FFP). Scientific Report. 2015;5:1-14.
Crossref - Jill E, Clarridge III. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840-62.
Crossref - Abbas S, Senthilkumar R, Arjunan S. Isolation and molecular characterization of microorganisms producing novel antibiotics from soil sample. Eur J Experiment Biol. 2014;4:149-55.
- Abdel-Mohsien H, Sasaki T, Tada C, Nakai Y. Characterization and partial purification of a bacteriocin-like substance produced by thermophilic Bacillus licheniformis H1 isolated from cow manure compost. Animal Science Journal. 2011;82:340-351.
Crossref - Beric T, Stankovic S, Draganic V, Kojic M, Lozo J, Fira D. Novel antilisterial bacteriocin licheniocin 50.2 from Bacillus licheniformis VPS50.2 isolated from soil sample. J Appl Microbiol. 2013;116(3):502-510.
Crossref - Smitha S, Bhat S. Thermostable Bacteriocin BL8 from Bacillus licheniformis isolated from marine sediment J Appl Microbiol. 2012;114(3):688-694.
Crossref - Maldonado M, Corona J, Gordillo M, Navarro A. Isolation and partial characterization of antifungal metabolites produced by Bacillus sp. IBA 33. Curr Microbiol.2009;59:646-650.
Crossref - Ser H-L, Ab Mutalib N-S, Yin, W-F, Chan K-G, Goh B-H, Lee L-H. Evaluation of antioxidative and cytotoxic activities of Streptomyces pluripotens MUSC 137 isolated from mangrove soil in Malaysia. Front Microbiol. 2015;6:1398.
Crossref - Durai S, Vigneshwari L. Balamurugan K. Caenorhabditis elegans-based in vivo screening of bioactives from marine sponge-associated bacteria against Vibrio alginolyticus. J Appl Microbiol. 2013;115:1329-1342.
Crossref - Krishnamoorthy K, Subramaniam P. Phytochemical Profiling of Leaf, Stem, and Tuber Parts of Solena amplexicaulis (Lam.) Gandhi Using GC-MS. Int Sch Res Notices. 2014;2014:1-13.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.