ISSN: 0973-7510
E-ISSN: 2581-690X
Electronic devices such as mobile phones, Bluetooth devices, electronic tabs, laptops, earphones and biometrics have all become an element in both the personal and professional life of human race. These electronic devices are in intense contact with us every day which might result in contamination of these devices and increase in the microbial load. Nowadays, the devices have become multiple interfaces leading to increase in the bacterial load and also the heat generated by various users make the devices a suitable breeding ground for the pathogenic organisms. The study is aimed at isolating and identifying microorganisms from electronic devices of health care professionals in and around Chennai. The study group was separated into the use of electronic devices like mobile phones, biometrics, laptops and electronic tabs utilized by doctors, nurses, interns and lab technicians. The samples were collected aseptically using sterile cotton swabs and transported to laboratory where isolation and identification of microbes were done using standard microbial testing. Total of 75 samples were collected in which 100% of the total sample were contaminated with heavy load of microorganisms. The species identified from the electronic devices were Pseudomonas sp., Bacillus sp., Streptococcus sp., Protease sp in abundance which is associated with pathogenic infections. Antibiotic sensitivity showed resistance against commercially available antibiotics on testing. Awareness among the health care professionals on personal hygiene and safe usage of the devices away from contamination are the possible ways to stay away from infectious pathogens.
Electronic devices, mobile phones, healthcare professionals, nosocomial infections, antibiotic sensitivity
In the present era of technological development, electronic devices have become an essential requirement in every walk of human life and life stumbles in its absence. Mobile phones, bluetooth, electronic tabs, laptops, computers, earphones and biometrics are communication devices being used every day. Even though we more often carry these electronic devices in our bags and pockets, our face, hands and ears come in contact with these electronic devices when we communicate. Hence these devices act as a vector for transmission of pathogenic microorganisms from person to person. The numerous advantages that these devices possess allow one to fail to notice the hazard over health making them a potential carrier for the spreading of pathogenic microorganisms causing diseases.1
In the perspective of a health care professional, surrounding environment plays a very important role in the spread of pathogenic microorganisms. They are continuously exposed to many infectious prone areas like handling patient’s samples in the laboratories or during handling patients in various health care setups. The health care professionals are often in contact with pathogenic microorganisms that could be disseminated to these electronic devices during their contact. Nosocomial infection is any disease that is acquired by a patient during their medical care. In the contemporary era of antibiotics, nosocomial infections are a very serious health problem.2 During patient care, especially in intensive care units usage of mobile phones and bluetooth extends the risk of nosocomial infections in patients even though they do not come in direct contact with these devices.3,4
Studies have shown that combination of frequent handling and the heat generated by the electronic devices creates a prime breeding ground for all sorts of microorganisms that are normally found in our skin and environment. Previous studies have indicated that the presence of medically important bacteria like Staphylococcus aureus, Staphylococcus epidermidis, Methicillin-Resistant Staphylococcus aureus (MRSA), Pseudomonas aeruginosa, Micrococcus luteus, Escherichia coli, Bacillus subtilis and Alternaria sp., Aspergillus niger, Cladosporium, Penicillium, Aspergillus flavus and Aspergillus fumigates were the main bacterial and fungal isolates found associated with mobile phones.5
Apart from all these, personal hygiene could be a regarding factor when electronic devices are concerned. Usage of electronic devices when we are in restrooms, working in laboratories, public transports, gardening, dusting, and so on also creates high risk environment for microorganisms to access your personal devices as you are in direct contact with them. This also paves the way for user to encounter detrimental infections.6
Sample Size
The present study was approved by the Ethics Committee for Students Projects, Sri Ramachandra Institute of Higher Education and Research. A total of 75 samples were collected (n = 75). The inclusion criteria included students, technicians, interns, doctors and nurses from hospitals in and around Chennai. The exclusion criteria included patients and children below 18 years.
Sample Collection
Samples were collected aseptically with a sterile swab that was moistened with sterile normal saline (rubbing the sterile swab over the surface of electronic devices). The collected swabs were aseptically transferred to laboratory for processing. The questionnaire was developed and validated which consisted of salient questions related to the types of electronic devices in contact, daily usage of electronic devices, exposure to various environments, mostly encountered infections and frequency in cleaning of the electronic device by the medical practitioner and clinicians. Informed consent and questionnaire were obtained from the participants prior to sample collection.7
Isolation and Identification of microorganisms
Samples collected aseptically were inoculated into nutrient broth, incubated at 37°C for 24 hours, following which quantification of bacteria was performed by plate count method and samples were also inoculated into yeast mannitol broth at 25°C for 48 hours. The bacterial samples were further subjected to identification tests such as gram staining, motility test, biochemical analysis (IMViC test, catalase test) and antibiotic sensitivity test. Lactophenol cotton blue staining was performed to identify the fungal species.8,9
Biochemical tests
Gram negative bacilli were identified by performing a series of biochemical tests IMViC [Indole, Methyl Red, Voges Proskauer test and Citrate utilization test]. Indole test- Test bacterial culture was inoculated with tryptone broth and incubated for 48 hours at 37°C. Following incubation few drops of Kovac’s reagent was added to test tubes. The tubes were observed for the formation of cherry red ring on the top.
Methyl red (MR) test- Test bacterial culture was inoculated with glucose phosphate broth (MRVP media) and incubated at 37°C for 48 hours. Following incubation 3-4 drops of Methyl Red (MR) indicator was added to the test tubes. The tubes were observed for development of red color after addition of MR indicator.
Voges Proskauer (VP) test-Test bacterial culture was inoculated with glucose phosphate broth and incubated for 48 hours at 37°C. Following incubation 0.5ml of Barrit’s solution A (α-naphthol) and equal amount of Barrit’s B solution was added to the test tubes and shaken. The tubes were observed for development of red color after addition of Barrit’s solution A and B.
Citrate utilization test-Test bacterial culture was inoculated with the simmon citrate agar in slanted position and incubated for 24- 48 hours at 37°C. If the organism has the ability to utilize citrate, color changes from green to blue. Gram positive cocci were identified based on their reaction in catalase test.
Catalase test- Slide method
A small amount of bacterial colony was transferred to a clean, dry slide using a sterile loop. A drop of 3% H2O2 was placed on to the slide and mixed. The slide was observed for the evolution of oxygen bubbles.
Antibiotic Sensitivity
Well diffusion method
The antibiotic sensitivity testing was done for the isolates by well diffusion method. The culture isolates were spread over the Mueller-Hinton agar medium with a sterile cotton swab aseptically. The plates were allowed to dry for 5 minutes. A hole with a diameter of 6 to 8 mm was punched aseptically with a sterile cork borer, and a volume (10 µL) of the commercially available antibiotics Ciproflaxin, Ampicillin, Levofloxacin and Amoxicillin were tested against the isolated gram positive and gram-negative organisms respectively. The plates were incubated after exposure for 24 hours at 37°C. After incubation the plates were observed for zone of clearance to determine whether the tested bacterial species are resistant or sensitive to the commercial antibiotic.
Sample Collection
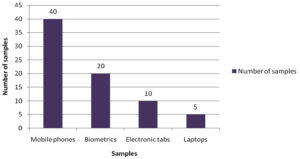
A total of 75 samples from the electronic devices were collected aseptically from various healthcare professionals and different healthcare setups. The distribution of sample collected from various sources of electronic devices is represented in (Fig.1) respectively.
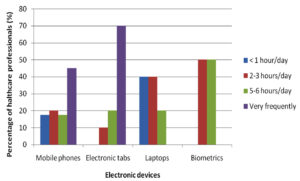
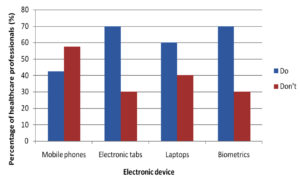
A questionnaire was given to every participant at the time of sample collection before which the questionnaire was validated as well. The questionnaire had included the usage of electronic devices on daily basis, which highlighted tabs being used more frequently in a day by 70% of people succeeded by mobile phones being used by 45% as represented in the (Fig. 2). Cleaning of electronic devices was a question of importance to the study showing that 60% of people cleaned their device surface while 40% did not as represented in (Fig. 3).
Quantification of microorganisms
Plate count agar method was used to quantify the number of microorganisms in different samples and was found that the number of microorganisms in each sample isolated were high.
Identification of microbes
Gram staining and lactophenol cotton blue staining techniques were performed for all the isolated bacterial and fungal isolates. The gram positive and gram-negative bacteria were determined respectively. The gram negative and gram-positive organisms were further classified and identified by biochemical analysis.
Selective media (MacConkey agar)
The samples were streaked onto MacConkey agar plates. The collected samples showed the presence of either lactose fermenting or non-lactose fermenting bacteria. Lactose fermenting bacteria were identified based on the formation of pink colonies and the non-lactose fermenting bacteria were identified based on the formation of colorless colonies on the agar.
Biochemical analysis
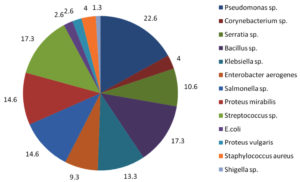
The samples were further subjected to biochemical analysis. The results of the indole, methyl red, voges proskauer, citrate and catalase test and the bacterial isolates that were identified at the species level is shown in (Table 1) and the (Fig.4) respectively.
Table (1):
Identification of isolated strains by gram staining and biochemical analysis.
| S.No | Samples | G +/- | M/NM | LF/NLF | I | MR | VP | Cit | Ca | Microorganism |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | Mobile phones | G- | + | NLF | – | – | – | + | + | Pseudomonas sp |
| G+ | – | NLF | – | – | + | + | + | Bacillus sp | ||
| G+ | – | NLF | – | + | – | – | + | Corynebacterium sp | ||
| G- | + | LF | – | + | – | + | + | Citrobacterfreundii | ||
| G- | + | LF | + | + | – | – | + | E.coli | ||
| 2. | Biometrics | G- | + | NLF | – | – | – | + | + | Pseudomonas sp |
| G- | + | NLF | – | – | + | + | + | Serratia sp | ||
| G+ | – | LF | – | + | – | + | – | Streptococcus sp | ||
| G+ | – | NLF | – | – | + | + | + | Bacillus sp | ||
| G- | + | LF | – | + | – | – | + | Salmonella sp | ||
| G- | + | NLF | – | + | – | + | + | Proteus sp | ||
| 3. | Laptops | G+ | + | NLF | – | + | – | – | + | Corynebacterium sp |
| G+ | – | NLF | – | – | + | + | + | Bacillus sp | ||
| G- | + | NLF | – | – | – | + | + | Pseudomonas sp | ||
| G- | + | NLF | – | – | + | + | + | Serratia sp | ||
| G+ | – | LF | – | + | – | + | – | Streptococcus sp | ||
| G- | + | LF | + | + | – | – | + | E.coli | ||
| G- | + | LF | – | + | – | – | + | Salmonella sp | ||
| 4. | Electronic tabs | G- | + | NLF | – | – | – | + | + | Pseudomonas sp |
| G+ | – | NLF | – | – | + | + | + | Bacillus sp | ||
| G+ | – | NLF | – | + | – | – | + | Corynebacterium sp | ||
| G- | + | LF | – | + | – | – | + | Salmonella sp | ||
| G- | – | NLF | +/- | + | – | + | + | Shigella sp |
* M- (+) Motile, (-) Non motile, LF- Lactose fermenting, NLF- Non lactose fermenting, analysis of selective media, Biochemical analysis Indole, Methyl red, Voges Proskauer, Citrate and catalase test (IMViC & Ca).
Fig. 4. Percentage of bacterial isolates found in the samples.
Antibiotic Sensitivity
Antibiotic sensitivity test was performed using the well diffusion method and zone of clearance was observed for all the bacterial isolates against the commercially available antibiotics.
Electronic devices such as mobile phones, laptops, computers, earphones and biometrics have become indispensable from the population of the present era. Among these mobile phones, tabs, laptops and biometrics are the most frequently used electronic devices. Studies show that mobile phones have emerged from being a mere communicable device among public to multipurpose, non-medical device used in the healthcare facility and general community.10 It is also due to its propinquity to the body that it is a vulnerable infection causing amenity. In the present study, 75 samples were aseptically collected from electronic devices like mobile phones, tabs, laptops and biometrics as represented in (Fig. 1) using sterile swabs that were moistened with normal saline. The data collected by the questioner from the participant clearly indicated that about 70% of the participants were in frequent contact with the electronic devices like mobile phones, tabs and ear buds. Majority of the participants never cleaned their electronic devices on regular basis, only 40% of the participants maintained their devices as represented in the (Fig. 2 and 3). The samples were collected from SRIHER, Xcellent Care Hospital and Anderson diagnostics & labs and transferred to laboratory for processing. Furthermore, the samples were processed by various microbial tests for isolation of bacteria and fungi respectively. Quantification of the microorganisms present in the samples was carried out with the help of plate count agar. It was observed that invariably all the 75 samples had loads of bacterial and fungal isolates which were more in number.
Identification of bacterial isolates was performed using gram staining, motility test, selective media and biochemical analysis. Both the gram positive and negative cocci and bacilli were identified. Motility test exhibited some samples with motile bacteria and others with non motile bacteria. A preliminary screening using differential media (MacConkey agar) indicated the presence of lactose fermenting and non-fermenting bacteria. Confirmatory biochemical test (IMViC and catalase test) had shown the bacterial species in particular which is represented in (Table 1) that demonstrates a comprehensive record of the isolates from the electronic devices.
Among the bacterial isolates Pseudomonas sp. was found to be the highest (22.6%), following which were Bacillus sp. (17.3%), Streptococcus sp. (17.3%), Salmonella sp. (14.6%), Proteus sp. (14.6%) and Shigella sp. being the least (1.3%) (Fig. 4). All the samples were sensitive to the commercially available antibiotics. Lactophenol cotton blue staining was performed for all the samples to identify the fungal species and based on the morphology; Aspergillus sp. and Candida sp. were identified.
This study revealed that the contamination of electronic devices of health care participants was mainly due to frequent handling during patient care. Nosocomial infections were reported in participants using mobile phones and tabs which were also seen in similar studies in hospital environment. This could be a potential risk factor for nosocomial infections to both health care professionals and people who come in contact with them.11,12 Bacterial and fungal infections are frequently been reported for COVID-19 affected patients. A recent study on 140 COVID-19 affected patients who acquired both bacterial and fungal infection after their admission in ICU with indication of infection by both the gram-positive, gram-negative and fungal organisms were subjected to be evident.13 Health care participants who did not regularly clean (disinfect) their mobile phone had higher bacterial contamination than those who regularly cleaned their mobile phone. This could be a probable area of future perspective where the general population along with the healthcare professionals could be advised on personal hygiene, cleaning their electronic devices, minimizing the use of devices in areas they are susceptible to encounter infectious pathogens or placing the gadgets in safe and sealed units free of contamination at their workplace. An alternate to this could be coating of the surface of electronic devices with antibiotics in the form of tempered films or protective coats. In the current scenario of COVID pandemic where immunity is at stake, risk of such secondary infections possesses great risk.
The present study showed elevated incidence of bacterial and fungal contamination from the electronic devices used by the health care participants. The results clearly indicated the significant load of bacterial and fungal contamination of microbial isolates isolated from the devices. As far as healthcare professionals are concerned, they are at high risk of encountering nosocomial infections or hospital acquired infections. These infections pose a serious threat to the lives of healthcare professionals as they are capable of developing serious diseases. A standard operating procedure on awareness of personal hygiene, cleanliness of devices, avoiding the usage of electronic devices in infection prone areas must be strictly followed in a healthcare sector.
ACKNOWLEDGMENTS
The authors would like to thank the Department of Biomedical Sciences, Sri Ramachandra Institute of Higher Education and Research for the necessities and facilities provided for the conduct of the study. We would also like to thank the Department of Microbiology, Sri Ramachandra Institute of Higher Education and Technology (DU), Porur, Chennai for providing us with the selective media for isolation of bacterial strains.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
- Al-Abdalall AHA. Isolation and identification of microbes associated with mobile phones in Dammam in eastern Saudi Arabia. J Family Community Med. 2010;17(1):11-14.
Crossref - Deepak KV, Alemu B, Dantse D, et al. Isolation And Characterization Of BacteriaFrom Mobile Phones Of Students And Employees At University Of Gondar, Ethiopia. Bulletin of Pharmaceutical Research. 2015;5(3):96-100.
- Suganya S, Judia H, Sumathy V. Isolation and identification of bacteria from covered and uncovered mobile phones. Int J Environ Sci. 2012;3(1):44-54.
Crossref - Bereket W, Hemalatha K, Getenet B, et al. Update on bacterial nosocomial infections. Eur Rev Med Pharmacol Sci. 2012;16(8):1039-1044.
- Dave S, Shende K. Isolation and Identification of Microbes Associated With Mobile Phones in Drug District in Chhattisgarh Region, India. J Environ Sci Toxicol Food Technol. 2015;1(6):71-73.
- Canales MB, Craig G, Boyd Jr. J, Markovic M, Chmielewski RA. Dissemination of Pathogens by Mobile Phones in a Single Hospital. Reconstructive Review. 2017;7(3):44-47.
Crossref - Zaman RMQ, Helmi NRM. Isolation of bacteria from mobile phones before and after decontamination : Study carried out at King Abdulaziz University, Jeddah, Saudi Arabia. Afr J Microbiol Res. 2017;11(35):1371-1378.
Crossref - Kotris I, Drenjancevic D, Talapko J, Bukovski S. Identification of microorganisms on mobile phones of intensive care unit health care workers and medical students in the tertiary hospital. Medicinski Glasnik. 2017;14(1):85-90.
Crossref - Rottier BP. Stain technology. Biotechnical & Histochemistry. 1954;29(6):225-239.
Crossref - Raghavendra RM, Sowmya GS, Rashmi PM, Sumana MN. Study of bacterial flora associated with mobile phones of healthcare workers and non-healthcare workers. Iran J Microbiol. 2017;9(3):143-51.
- Sepehri G, Talebizadeh N, Mirzazadeh A, Mir-Shekari TR, Sepehri E. Bacterial contamination and resistance to commonly used antimicrobials of healthcare workers’ mobile-phones in teaching hospitals, Kerman, Iran. Am J Appl Sci. 2009;6(5):806-810.
Crossref - Ulger F, Esen S, Dilek A, Yanik K, Gunaydin M, Leblebicioglu H. Are we aware how contaminated our mobile phones with nosocomial pathogens. Ann Clin Microbiol Antimicrob. 2009;8:4-7.
Crossref - Bardi T, Pintado V, Gomez-Rojo M, et al. Nosocomial infections associated to COVID-19 in the intensive care unit: clinical characteristics and outcome. Eur J Clin Microbiol Infect Dis. 2021;40(3):495-502.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.