ISSN: 0973-7510
E-ISSN: 2581-690X
In the present world of industrial revolution, spills of petroleum products and oil are one of the major sources of the contamination in the ecosystem and cause serious health hazards for humans and livestock. Therefore, the removal of these pollutants has become necessity of the time. Applications of new techniques such as bioremediation and biodegradation, over conventional methods are much more promising for safe and sustainable environment. Aim-The aim of this study is to isolate and characterize microbial strains from effluent of Mathura Refinery – Indian Oil Corporation Limited, the microscopic and biochemical studies and their potential of bioremediation. Bacteria were isolated from Mathura refinery IOCL, Based on the biochemical studies six morphologically distinct bacterial strains with promising bioremediation role were identified. To check the degradation of crude oil sample by bacterial isolates Bushnallhass media and separation funnel method were performed. Six bacteria were isolates from Mathura refinery effluentsand all six bacteria have ability to degrade the crude oil in different range. Maximum potential to degrade the crude oil were identified in Pseudomonas. This study might be important in an important step towards bioremediation techniques.
Biodegradation, Bioremediation, Petroleum products, Safe and sustainable environment.
Water pollution is one of the serious ecological threats among various types of pollutions. Various anthropogenic activities, such as heavy industrialization including crude oil refinery, chemical, textiles, nuclear industries etc., have led to the contamination of ground water in both urban and rural area leading to serious health hazard1.
Release of crude oil or petroleum products in the ecosystem severely affects the biotic communities. To overcome such ecological extermination several physical and chemical methods are conventionally used whichleads to the conversion of comparatively less toxic forms instead of the complete degradation and removal2. However, the modified form of petrochemical still causes cellular toxicity leading to various deformities. The hydrocarbons released in the ecosystem can be metabolized by many communities of bacteria inbiodegradable nontoxic products3-4. The advantage of bacteria mediated remediation of ecosystem (i.e. bioremediation) over conventional methods is that the bioremediation provides a complete solution of toxic substances rather than converting their forms in a cost effective manner.
Use of microbial strainsfor treatment of polycyclic aromatic hydrocarbons from contaminated locations has already been reported1,6,9.
Numerous residential microflora naturally do have inherent properties for degrading these pollutants from soil and aquatic ecosystems. The purpose of present work is isolation and characterization of bacterial strains from oil refinery effluent and their identification through colony morphology, microscopic and biochemical studies6-7.
Toxic pollutants cause harm to living and non-living things but before devising solutions to cure them, we need to understand that their sources, leaching process, chemical conservation and their mode of deposition8.
Sample collection
Industrial effluent was obtained, in sterile sample collection screw cap bottles of 100 ml volume aseptically, from the effluents of oil petroleum refinery, Mathura (IOCL Mathura Refinery Mathura, Uttar Pradesh, India). The collected sample was then immediately bought to the laboratory for further processing.
Isolation of pure bacterial strains
Initial isolation
For initial isolation of bacterial strains serial dilution in 0.8% NaCl solution of collected industrial effluents was performed. 5ml NaCl solution was transferred in five test tubes labeled as 10-1 to 10-5. Followed by addition of 50לl inoculum (oil refinery effluent) in the first tube and serial diluted. From 10-1, 10-3 and 10-5 tubes 10לl sample was spread on solidified Nutrient Agar plates and incubated overnight at 37°C.
Primary bacterial screening
After overnight incubation colonies found on Nutrient medium plates, were selected for secondary screening of pure culture. The single morphological bacterial colony was transferred separately to another Nutrient Agar Medium (NAM) plates by the means of inoculation loop and were incubated overnight at 37°C. The isolated bacterial strains were grown-up on MacConkey Agar media with pH 7.2 at 37°C. Finally pure and axenic cells were transferred aseptically to NAM slants and test tubes containing broth media for further investigation in incubator at 37°C3.
Confirmatory test for oil degradation microbes by using bushnellhaas media
For detecting oil degrading ability of axenic strains, method introduced by Bushnell and Haas, (1941), has been used with Bushnell-Haas media. Bushnell Haas Agar was used for microbiological examination of crude oil it uses for the study of microbial utilization of hydrocarbon. This media contains Composition (g/l) Magnesium Sulphate 0.2, Calcium Chloride 0.02, Monopotassium Phosphate 1.0, Dipotassium Phosphate 1.0, Ammonium Nitrae 1.0, Ferric Chloride 0.05, Agar 20.0, Final pH (at 25°C) 7.0±0.2.
After preparation of Bushnell Haas agar plate the isolated bacterial cultures were inoculated into the medium3 and one control plate and six bacteria isolates with Bushnell Hass Agra with the inoculum of six bacterial isolates and 15 ml of crude oil sample in each plates were kept inthe incubation at 37°C for 7 days. Finally positive results were observed by measuring the quantity of oil degraded by the each bacterial strain after 7 days.
Inoculation of pure culture isolates into oil containing nutrient broth
In this stage the oil degradation capacity is confirmed by each bacterium. 20 ml of crude oil sample were taken in six conical flask of nutrient broth. Place the conical flask in autoclave at 15lb on 120°C for 30 minutes. Inoculate the pure culture isolates of bacteria with the help of inoculation loop under Laminar in each flask. Put the flasks for incubation at 37°C in incubator for 30 days, with the interval of 3-5 days put the conical flasks in the shaker also to provide the proper agitation of bacteria.
Separation funnel method for measurements of oil degradation analysis
After the incubation of 30 days degraded oil amount were measured with the help of separation funnel method by putting the nutrient broth with oil and bacteria isolates in separation funnel for 35 minutes. Amount of oil measured in measuring cylinder with the help of micropipette for each sample and analysis shows that all six bacteria have ability to degrade the amount of crude oil sample after measuring the quantity of crude oil.
Biochemical analysis
For biochemical analysis, we used VITEK2 which is an automated system for microbial growth analysis and provide highly accurate data. It performs 64 individual tests in 64 different well like various metabolic activities (alkalinization, acidification, enzymes hydrolysis and growth) in the presence of inhibitory substance.
To identify the different classes of organism, four reagents card were used; turbidity was also measured with the help of turbidity meter- DensiChek.
Table (1):
The bacterial class identification and suspension turbidity used for card inoculation.
S. No. |
Bacteria classes |
Identification |
McFarland Turbidity |
|---|---|---|---|
1 |
GN |
Gram negative fermenting and non-fermenting bacilli |
0.50-0.63 |
2 |
GP |
Gram positive cocci and non-spore forming bacilli |
0.50-0.63 |
3 |
YST |
Yeast and yeast like organisms |
1.80-2.20 |
4 |
BCL |
Gram- positive spore- forming bacilli |
1.80-2.20 |
Isolation of bacteria
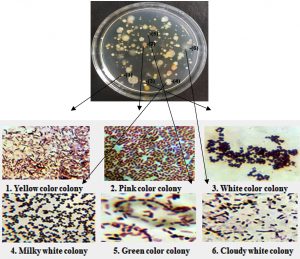
A total of six bacteria from refinery effluents were screened on Nutrient agar media. By understanding the biochemical and morphological analysis six strains are Bacillus sp1,Algoriphagus, Bacillus sp2, Acinetobacter, Pseudomonas, Bacillus sp3.
Identification and characterization of oil degrading bacterial isolates
On the basis of colony characterization, morphology, species, growth, stain test, colony size, colony form (Table 2) and biochemical analysis (Table 3) six different bacteria were identified. From the analysis one bacteria out of six were gram positive remaining five were gram negative; Three out of six bacteria were different shades of white in color.
Table (2):
Morphological characteristics of the bacterial isolates cultured on Nutrient agar medium.
S.No |
Colony Color |
Microscopic View |
Genus |
Gram Staining |
Colony Size |
Colony Form |
|---|---|---|---|---|---|---|
1 |
Yellow color colony |
Bacillus |
Gram Positive |
Small |
Circular |
|
2 |
Pink color colony |
Algoriphagus |
Gram Negative |
Medium |
Irregular |
|
3 |
White color colony |
Bacillus sp. |
Gram Negative |
Small |
Irregular |
|
4 |
Milky white colony |
Acinetobacter |
Gram Negative |
Large |
Irregular |
|
5 |
Green color colony |
Pseudomonas sp |
Gram Negative |
Small |
Circular |
|
6 |
Cloudy white colony |
Bacillus sp |
Gram Negative |
Medium |
Irregular |
Table (3):
Biochemical Test Results.
| GRAM NEGATIVE | GRAM POSITIVE | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Well | Test | Mnemonic | Algoriphagus | Bacillus sp. | Pseudomonas sp | Bacillus sp | Well | Test | Mnemonic | Bacillus | |
| 2 | Ala-Phe-Pro ARYLAMIDASE | APPA | – | – | (+) | – | 2 | D-AMYGDALIN | AMY | – | |
| 3 | ADONITOL | ADO | – | – | – | – | 4 | PHOSPHATIDYLINOSITOL PHOSPHOLIPASE C | PIPLC | – | |
| 4 | L-Pyrrolidonyl- ARYLAMIDASE | PyrA | – | (+) | (+) | – | 5 | D-XLOSE | dXYL | – | |
| 5 | L-ARABITOL | IARL | – | – | – | – | 8 | ARGININE DIHYDROLASE 1 | ADH1 | – | |
| 7 | D-CELLOBIOSE | dCEL | – | – | – | (+) | 9 | BETA- GALACTOSIDASE | BGAL | + | |
| 9 | BETA- GALACTOSIDASE | BGAL | – | – | – | – | 11 | ALPHA-GLUCOSIDASE | AGLU | (+) | |
| 10 | H2S PRODUCTION | H2S | – | (+) | – | – | 13 | Ala-Phe-Pro ARYLAMIDASE | APPA | + | |
| 11 | BETA-N-ACETYL GLUCOSAMINIDASE | BNAG | – | (+) | – | – | 14 | CYCLODEXTRIN | CDEX | – | |
| 12 | GlutamylArylamidasepNA | AGLTp | – | – | – | – | 15 | L-Asppartate ARYLAMIDASE | AspA | + | |
| 13 | D-GLUCOSE | dGLU | + | (+) | – | (+) | 16 | BETA GALACTOPYRANOSIDASE | BGAR | + | |
| 14 | GAMMA-GLUTAMYL-TRANSFERASE | GGT | – | – | (+) | – | 17 | ALPHA-MANNOSIDASE | AMAN | – | |
| 15 | FERMENTATION/GLUCOSE | OFF | – | – | – | – | 19 | PHOSPHATASE | PHOS | + | |
| 17 | BETA GLUCOSIDASE | BGLU | + | (+) | – | (+) | 20 | Leucine ARYLAMIDASE | LeuA | + | |
| 18 | D-MALTOSE | dMAL | – | (+) | – | – | 23 | L-Proline-ARYLAMIDASE | ProA | + | |
| 19 | D- MANNITOL | dMAN | – | – | – | (+) | 24 | BETA-GLUCURONIDASE | BGURr | + | |
| 20 | D-MANNOSE | dMNE | – | – | – | (+) | 25 | ALPHA-GALACTOSIDASE | AGAL | + | |
| 21 | BETA- XYLOSIDASE | BXYL | – | – | – | – | 26 | L-Pyrrolidonyl-ARYLAMIDASE | PyrA | + | |
| 22 | BETA-ALANINE arylamidasepNA | BALap | – | – | – | – | 27 | BETA-GLUCURONIDASE | BGUR | – | |
| 23 | L-Proline ARYLAMIDASE | ProA | + | – | (+) | – | 28 | Alanine ARYLAMIDASE | AlaA | – | |
| 26 | LIPASE | LIP | + | – | (+) | – | 29 | Tyrosine ARYLAMIDASE | TyrA | + | |
| 27 | PALATINOSE | PLE | – | – | – | – | 30 | D-GALACTOSE | dSOR | – | |
| 29 | Tyrosine ARYLAMIDASE | TyrA | + | – | (+) | – | 31 | UREASE | URE | – | |
| 31 | UREASE | URE | – | (+) | – | – | 32 | POLYMIXIN B RESISTANCE | POLYB | – | |
| 32 | D-SORBITOL | dSOR | – | – | – | (+) | 37 | D-GALACTOSE | dGAL | – | |
| 33 | SACCHAROSE/SUCROSE | SAC | – | – | – | (+) | 38 | D-RIBOSE | dRIB | – | |
| 34 | D-TAGATOSE | dTAG | – | – | – | (+) | 39 | L-LACTATE alkalinization | ILATk | – | |
| 35 | D-TREHALOSE | dTRE | – | (+) | – | (+) | 42 | LACTOSE | LAC | – | |
| 36 | CITRATE(SODIUM) | CIT | + | – | (+) | – | 44 | N-ACETYL-D-GLUCOSAMINE | NAG | – | |
| 37 | MALONATE | MNT | – | – | – | – | 45 | D-MALTOSE | dMAL | – | |
| 39 | 5-KETO-D-GLUCONATE | 5KG | – | – | – | – | 46 | BACITRACIN RESISTANCE | BACI | – | |
| 40 | L- LACTATE Alkalinization | ILATk | – | (+) | (+) | – | 47 | NOVOBIOCIN RESISTANCE | NOVO | – | |
| 41 | ALPHA- GLUCOSIDASE | AGLU | – | (+) | – | – | 50 | GROWTH IN 6.5% NaCl | NC6.5 | – | |
| 42 | SUCCINATE alkalinization | SUCT | + | – | (+) | – | 52 | D-MANNITOL | dMAN | – | |
| 43 | BETA-N-ACETYL-GALACTOSAMINIDASE | NAGA | – | – | – | – | 53 | D-MANNOSE | dMNE | – | |
| 44 | ALPHA-GALACTOSIDASE | AGAL | – | – | – | – | 54 | METHYL-B-D-GLUCOPYRANOSIDE | MBdG | – | |
| 45 | PHOSPHATASE | PHOS | + | (+) | – | – | 56 | PULLULAN | PUL | – | |
| 46 | Glycine ARYLAMIDASE | GlyA | – | – | (+) | – | 57 | D-RAFFINOSE | dRAF | – | |
| 47 | ORNITHINE DECARBOXYLASE | ODC | – | – | – | – | 58 | O/129 RESISTANCE(comp.vibrio.) | O129R | – | |
| 48 | LYSINE DECARBOXYLASE | LDC | – | (+) | – | – | 59 | SALICIN | SAL | – | |
| 53 | L-HISTIDINE Assimilation | IHISa | – | – | – | – | 60 | SACCHAROSE/SUCROSE | SAC | – | |
| 56 | COUMARATE | CMT | + | (+) | – | – | 62 | D-TREHALOSE | dTRE | – | |
| 57 | BETA-GLUCORONIDASE | BGUR | – | – | – | – | 63 | ARGININE DIHYDROLASE | ADH2s | – | |
| 58 | O/129 RESISTANCE (comp.vibrio) | O129R | – | (+) | – | – | 64 | OPTOCHIN RESISTANCE | OPTO | – | |
| 59 | Glu-Gly-Arg-ARYLAMIDASE | GGAA | – | – | – | – | |||||
| 61 | L-MALATE Assimilation | IMLTa | + | – | (+) | – | |||||
| 62 | ELLMAN | ELLM | – | – | (+) | – | |||||
| 64 | L-LACTATE assimilation | IIATA | – | – | (+) | – | |||||
Screening of bacteria on crude oil
Crude oil has different hydrocarbons it also includes olefins paraffin and aromatic compounds10,11. Crude oil is the great substrate for screening of hydrocarbon degradation capabilities in microorganism because it has the variation in hydrocarbons and its composition3,4.
The ability of oil degradation was determine by the growth of bacteria on Bushnell Haas Agar media and add the same amount of oil in all six petriplates and keeps it for up to 7 days at 30°C. The effect of variation in quantity of oil after seven days defined each bacterium has the ability of oil degradation at different level [3].Separation funnel method also shows that the amount that remains after analysis was less than the amount that added at the initial, after considering the amount remains in control in both the analysis. This ability determines that all 6 isolates obtained from the sampling site have metabolic active and ability to utilize the hydrocarbon as their energy source
Identification of potent bacterial strain for oil degradation
After Bushnell Hass media and separation funnel analysis, the confirmatory test for oil degradation was found maximum in Pseudomonas12 by consuming the maximum oil amount in both the analysis. Other than Pseudomonas, bacillus sp1, Algoriphagus, Bacillus sp2, Acinetobacter and Bacillus sp3 also have potential of crude oil degradation.
Nowadays, activities related to the petrochemical industry are one of the serious environmental concerns in terms of releases of hydrocarbon contaminates in the ecosystem13. These are the organic toxic pollutants often associated with neurotoxicand carcinogenic activities in humans. Conventional methods for disposal of these pollutants have their limits in terms of effectiveness and cost. Oil degradation bacteria are present in extensive range in environment and well known for their ability of degradation of hydrocarbons present in oil14. Microorganism benefits in the removal of contaminant because of the presence of some special proteins and enzymes.
Application of nonpathogenic microbial stains for solving this problem is a promising tool for treatment of contaminated sites9. These microorganisms utilize complex toxic hydrocarbon contaminants into their metabolic cycles and transform them into simple non toxic forms with complete mineralization. Various physical and chemical factors are responsible for influencing the phenomena. Numerous residential microfloraas naturally possess inherent capabilities of degrading these toxic pollutants from soil and water ecosystems.
Hydrocarbon degrading potential of pseudomonas has recently been delineated and attributed to presence of naphthalene dioxygenase operon in its genome which is involved in the degradation of Benzene, Toluene, Ethylbenzene, Xylene15. Similarly biodegradation of patroleum hydrocarbon has also been reported by Bacillus sp.16 a metagenomic study has also reported the enrichment of Acinetobacter sp. in a marine environment after an oil spill event17. All these recent works corroborate with our findings.
The successful bioremediation techniques and approaches practiced using microbes for petroleum hydrocarbon degradation3. The road ahead for implementation of these organisms for the improvement of the environment and ultimately, public health is indeed long and worthwhile.
The aim of this study was to proposes the ecofriendly approach for the problem of discharging waste in the environment. We further endorse our approach for identifying native bacterial strains from refinery effluent, for consumption of used engine oil.
Six bacterial strains were isolated, purified and identified on the basis of colony characteristic, microscopic studied and biochemical analysis. In which oil degradation capacity was found in Pseudomonas bacillus sp1, Algoriphagus, Bacillus sp2, Acinetobacter and Bacillus sp3. Oil degrading microorganisms are wide spread in nature. They are well known for their ability to degrade variety of hydrocarbons present in the crude oil and harbor catabolic enzymatic activity to utilize organic contaminants as sole carbon and energy source converting them into less harmful substances12. Use of indigenous microorganisms aids in the removal of organic contaminants due to the specific enzyme systems synthesized by microorganisms.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Patel NK, Pethapara H., Isolation and screening of hydrocarbon degrading bacteria from soil near Kadi (Gujarat) region. International Journal of Research in BioSciences, 2013; 2(4): 10-16.
- Das N, Chandran P., Microbial Degradation of Petroleum Hydrocarbon Contaminants: An Overview, SAGE-Hindawi Access to Research, Biotechnology Research International, 2011; 2011(941810).
Crossref - Bushnell L.D., Haas H. F. The Utilization of Certain Hydrocarbons by Microorganisms. Journal of Bacteriol, 1941; 41(5): 653–673.
- Farag M., Shaieb, Abdelhamid H.,Ahmed Issa., Studies on crude oil degrading Bacteria isolated from Libyan Desert. International journal of current microbiology and applied science, 2015; 4(2): 920-927.
- Shahab S, Shafi I, and Shafi N. Indigenous Oil Degrading Bacteria: Isolation, Screening and Charac-terization. National Journal of Health Sciences, 2017; 2: 100-105.
Crossref - Sagheer A, Dobhal S, Tomar V. A Comparative Study of Oil Degradation with Used and Unused Engine Oil by Microbes Isolated From Water Sample of Mechanic Workshops. Agriculture research & technology open access journal, 2016; 10(2).
Crossref - Duruibe ,MOC Ogwuegbu, Egwurugwu JN. Heavy metal pollution and biotoxic effect. International Journal Physics Science, 2007; 2: 112–118.
- Han Y, Ren JN, Cai TT, Li GC, Zhang H. Isolation of Oil-degrading Bacteria and Treatment of Oil Wastewater. IOP Conference series. Earth and Environmental Science. IOP Conference Series: Earth and Environmental Science, 2018; 199(2): 1755-1315.
Crossref - Pantoja D.P., Gonzalez B., Pieper D.H., Aerobic Degradation of Aromatic Hydrocarbons. Handbook of Hydrocarbon and Lipid Microbiology Springer link, 2016; 1-44.
Crossref - Udgire M, Shah N, Jadhav M. Enrichment Isolation and Identification of Hydrocarbon Degrading Bacteria. International Journal Current Microbiology Application Science, 2015; 4(6): 708-13.
- Nogales J., Garcia J L, Doaz E. Degradation of Aromatic Compounds in Pseudomonas. A Systems Biology View. Aerobic Utilization of Hydrocarbons, Oils and Lipidsspringer link, 2017; 1-49.
Crossref - Pratiwi and Titah H.S. Isolation and Screening of Diesel-Degrading Bacteria from the Diesel Contaminated Sea water at Kenjeran Beach. Environmental Asia, 2016; 9(2) :165-169.
- Macaulay, B.M., Rees D. Bioremediation of oil spills: a review of challenges for research advancement. Annals of Environmental Science, 2014; 8: 9-37.
- V Imperato, M Portillo-Estrada, BM Mcammond, Y Douwen, JDV Hamme, SW Gawronski, et al. Genomic Diversity of Two Hydrocarbon-Degrading and Plant Growth-Promoting Pseudomonas Species Isolated from the Oil Field of Bobrka (Poland). Genes (basels), 2019; 10(6): 443.
Crossref - D Wang, J Lin, J Lin, W Wang, S Li. Biodegradation of Petroleum Hydrocarbons by Bacillus subtilis BL-27, a Strain with Weak Hydrophobicity. Molecules, 2019; 24(17): 3021.
Crossref - CS Neethu, C Saravanakumar, R Purvaja, RS Robin, R Ramesh. Oil-Spill Triggered Shift in Indigenous Microbial Structure and Functional Dynamics in Different Marine Environmental Matrices. Scientific Reports, 2019; 9(1).
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.