ISSN: 0973-7510
E-ISSN: 2581-690X
Algae have received global attention as a renewable resource of biodiesel and may play an important role as a component contributing to the economic growth of the country. A variety of blue-green and green algae are found in the wetlands of various regions of India, however, they have hitherto remained untapped. Present research work deals with the isolation of gene responsible for expression of delta 9 acyl-lipid desaturase from Nostoc sp. which is a potent source of lipids that can be further utilized in biodiesel production. Due to the ever increasing fuel need it is of great importance to find an alternative feedstock for energy source. The above mentioned gene was then sequence characterized. The ORF sequence was identified in the gene sequence of Acyl lipid desaturases from Nostoc sp. Eight similar sequences were also identified which are present in different organism. The ORF sequence belongs to the Functional domain (Membrane_FADS) –like superfamily. Four different motifs of different length were also identified.
9 acyl-lipid desaturase, Nostoc sp., biodiesel.
The world is about to face the predicament in energy sources. The consumption of fossil fuels and the available energy sources are the main problems needed to be solved. This critical situation that the world is about to face is a threat to modern human existence. Energy crisis will arise when there will not be enough energy sources to survive the demands of the world as the demand for energy increases. The focus is on the biomass energy and there are biochemical processes for conversion of biomass into biological hydrogen gas, biodiesel and ethanol which are the possible biofuels of future generation. Algae are simple autotrophic organisms and from simple inorganic molecules such as carbon dioxide they produce complex organic compounds using energy from light or inorganic chemical reactions. These complex organic compounds include a significant amount of triacylglycerol which can be utilized to produce biodiesel using the process known as Transesterification. Thousands of forms of algae have been identified, but some algae strains are more suitable for biofuel production because of their high oil yield. Genetically modified strains of algae are being developed for algae biofuels for yielding high lipid-content algae. Some companies have developed algae strains with unique characteristics for example, strains of algae that produces ethanol at a rate of over 6,000 gallons per acre per year and genetically modified algae that grows in the dark. This is only possible if we identify the genes responsible for lipid production in algae. Oil producing(Oleaginous) microalgae like Nostoc sp. are good sources of lipids for the production of biodiesel. Long-chain polyunsaturated fatty acids (PUFA) synthesis pathways are not very well known in Nostoc sp and they affect the production of biodiesel. Acyl lipid desaturases are fundamental enzymes in PUFA synthesis pathway. These enzymes convert a saturated bond into an unsaturated bond between carbon molecules in fatty acids and they are further esterified to glycerolipids. (Sing et al., 2011).
In the present research work gene responsible for the expression of delta 9 acyl-lipid desaturase of Nostoc sp. was isolated and its sequence characterization was performed. Delta 9 acyl-lipid desaturase is known for converting saturated fatty acids into unsaturated fatty acids that is there are involved in the production of unsaturated fatty acids.The ORF sequence was identified in the gene sequence of Acyl lipid desaturases from Nostoc Sp. Eight similar sequences were also identified which are present in different organisms. The ORF sequence belongs to the Functional domain (Membrane_FADS) – like superfamily. Four different motifs of different length were also identified.
The algal strains namely Nostoc sp., Vaucheria sp., Diatom sp., Microcystis sp. and Oedogonium sp. were screened for fast growth and high lipid content. Algal sp. which was the highest in lipid production was used for sequencing and genome analysis.
Isolation of algae DNA and Sequencing of lipid producing gene
Genomic DNA extraction from the algal sample was performed using the method given by Huang et al. (2000). For the DNA extraction the algae cells were harvested by centrifugation at 5000 rpm for 10 minutes. The pellet cells were frozen in liquid nitrogen prior to grinding in pre-chilled pestle and mortar. Powdered microalgal mass was immediately transferred to falcon tube containing 3ml CTAB buffer (pre-incubated at 600C) and was incubated in water bath at 600C for 1 hour with intermittent shaking of the tube to allow the DNA to be released from the cells. Then, an equal volume (3ml) of chloroform:isoamylalcohol (24:1) was added to the tube. The tube was slightly shaken and centrifuged at 10,000 rpm for 10 minutes at 400C. The supernatant was collected in a fresh tube and double the volume of chilled isopropanol was added to the tube. The tube was again centrifuged at 14,000 rpm for 10 minutes at 40C. The supernatant was carefully removed and equal amount of chilled 70% ethanol was added to the tube and centrifuged at 14,000 rpm for 5 minutes. Finally, ethanol was carefully removed and the pellet was air dried in the laminar flow hood. The pellets were suspended in 100µl TE buffer (10mM Tric-HCl, pH 8.0; 1mM EDTA, pH 8.0) and stored at 40C.
Quantification of Isolated DNA
he quantity of the isolated DNA was checked in UV-VIS spectrophotomer (Vivaspec Biophotometer, Germany). From the stock 1¼l DNA was mixed with 49-¼l sterile distilled water to get 50 times dilution. DNA concentration was determined by the equation:
1 OD260 unit = 50 µg/ml (1 ng/µl =1 µg/ml).
The A260/A280 ratio was recorded to check the purity of DNA preparation. (Desjardins and Conklin, 2010)
Primer Designing and synthesis
The gene selected for the primer designing was delta 9 acyl-lipid desaturase with an amplicon size of 1324 bp. The specific primers were designed using Primer Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/) and the designed oligonucleotides were synthesized in Sigma Corporation USA.
PCR Amplification
For the amplification of DNA, PCR was used. Table 1 shows the reagents used in the PCR reaction mixture and Table 2 shows the temperature profile used for the PCR.
Table (1):
Reagents and the optimal PCR reaction mixture.
PCR components |
Volume (μl) |
|---|---|
Nuclease free water |
10.75 |
10X reaction buffer with MgCl2 (1.5mM) |
2.00 |
dNTP mix (2.5mM) |
2.00 |
Primer I (10picomoles/ μl) |
2.00 |
Primer II (10picomoles/ μl) |
2.00 |
Taq DNA polymerase (5U) |
0.25 |
Template DNA (50ng/ μl) |
1.00 |
Total volume |
20.0 |
Table (2):
PCR temperature profile.
Initial denaturation |
94°C for 2 min |
Denaturation |
94°C for 1min |
Annealing |
48°C for 30 s 30 cycles |
Extension |
72°C for 2min |
Final extension |
72°C for 6 min |
Genomics of the selected algae sample
Gene named, delta 9 acyl-lipid desaturase was nucleotide sequenced from the selected algal sample which was a good source of lipid for the production of biodiesel. Acyl lipid desaturases are fundamental enzymes in PUFA synthesis pathway. These enzymes convert a saturated bond into an unsaturated bond between carbon molecules in fatty acids and they are further esterified to glycerolipids.
Sequence analysis
Sequencing of delta 9 acyl-lipid desaturase gene was carried out by Sanger sequencing method (Sanger and Coulson, 1975) using ABI automated DNA sequencer. Following sequence analysis tools were used:
- Open reading frame (ORF) was predicted by using ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
- The identification of predicted ORF protein is carried out by using BLASTP server (http://blast.ncbi.nlm.nih.gov/Blast.cgi) at National Center for Biotechnology Information (NCBI). Five most identical sequences were identified.
- Sequence alignment and phylogenetic analysis was carried out by using ClustalW server (http://www.ebi.ac.uk/Tools/msa/clustalw2/) at European Bioinformatics Institute (EBI).
- The amino acid sequence of the selected enzymes were further used to identify the presence of conserve domain with the help of the conserve domain identification tool. The NCBI conserved domain database search was performed with NCBI conserved domain search program (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer et al).
- The search was performed at E-Value 0.01 and the database used for conserve domain is CDS search with tool cddv2.25. The motifs were predicted using motif search server at http://www.genome.jp/tools/motif/
The algal strains namely Nostoc sp., Vaucheria sp., Diatom sp., Microcystis sp. and Oedogonium sp. were screened to find the best candidate for lipid production and among the above five strains Nostoc sp. was the most potent strain for lipid production. Therefore Nostoc sp. was used for sequencing and further analysis.
Isolation of algae DNA and Sequencing of lipid producing gene
Genomic DNA isolated from the purified cultures of microalgae was run in 0.8% Agarose gel to verify the quality of the DNA. Figure 1 shows the PCR gel picture of the DNA sample with the ladder. Arrow in the figure signifies the size of the gene in comparison to the ladder i.e. 1300bp. comparison to the ladder i.e. 1300bp) Jin et al. (1997) tested 70 species of brown, red, and green algae for DNA quality using PCR. Hong et al. (1997) found that DNA extracted from most green algae by the CTAB method were of sufficient quality to be used as a template for PCR amplification.

Fig. 1. PCR gel picture of algal DNA (arrow signifies the size of the gene in comparison to the ladder i.e. 1300bp)
Quantification of Isolated DNA
The concentration of the isolated DNA was measured to be 325ng/ ¼l and OD ratio (A260/280) was estimated to be 1.82. This signified DNA to be pure with very minor RNA contamination.
Assessment of purity of DNA is an important step. The most commonly used assay is the A260/A280 ratio. DNA absorbs almost twice as much UV at 260 nm as it does at 280 nm. If there is no protein present, the target ratio is 1.8. However, because 280 nm is a peak for protein absorption, protein contamination will increase the A280 reading but have little effect on the A260 reading. Thus the A260/A280 ratio will be lower than 1.8. (Desjardins and Conklin, 2010).
Primers used
To isolate the gene delta 9 acyl-lipid desaturase PCR was run with the following primers:
Forward
(5’ CCTGAGAGA GGCAATT CA CC3’) and
Reverse primer (5’TGAGGGGGACTTGAAACAAC 3’).
Sequencing
Gene obtained as PCR product was sequenced. Sequence was 1324 bp long. The isolated delta 9 acyl-lipid desaturase nucleotide sequence in FASTA format is given below.
>gi Nostoc sp. 36 desC2 gene for delta 9 acyl-lipid desaturase, strain 36 C C TGAGAGAG GCAATTCACCTCAACTCAGGTGGATAAATG TGGTATTTTTTGGTGTATTCC ATGCCTTAGCACTTCTGTCTCCTT GGTTTTTCTCTTGGTCAGCATT AGGTTTACTAGTGTTTCTGC ACTGG TTATTCG GGAGCATTGGTAT TTGC TTGGGATATCAC GACTACTGAGCCATAAG AGTTTCCAAGTTCCTAAGTGGTTA GAGTATGCGATCGCCCTTA TTGGAGCGC TGGCTTTG CAAGGT GGGCCAATTTTTTGGGTA GGTGG ACACCGCCAGCATCACGC CCACACGGAAGATAT TGACCTAGAT CCCTATTCCGCCCAAAAAGGA TTTTGGTGGAGCCATATA CTATGGATTTTCTAC CCGCGCCCAGAATTTTTTG ACTATGACACCTATAAAAAATATGCTCCT GACTTAGCAAGA CAACCCTTTTAT TGCTGGCTGGATCGC TACTTCTTGCTGTTGC AAATACCCTTTGCG CTGCTGCTATACGTCTTA GGAGGATGGCCTTTCGTATTC TACGGAGTGTTTCTCAGATGTGTAC TGCTTTGGCACT CAACCTGG TTTGTGAACTCAGCATCACATCTG TGGGGTTATCGC ACCTTTGATGCTGATGATG GCGCTCGTA ATCTTTGGTGGGTATC CATCGTAACTTATGGA GAAGGATGGCACAATAACCAT CATACTTATC CCCACATGGC GAAATCTGGGTTGTTTTGGTGG GAGATTGATGTTACTT GGTGGAGCATCCAACTTTT GCAGACTTTAGGTTTAGCCA AAAAAGTTGTTTCAAGTCCCCCTCA
ORF detection
An ORF is a sequence of DNA that starts with start codon “ATG” (not always) and ends with any of the three termination codons (TAA, TAG, TGA). Depending on the starting point, there are six possible ways (three on forward strand and three on complementary strand) of translating any nucleotide sequence into amino acid sequence according to the genetic code .These are called reading frames. Eukaryotic gene finding is altogether a different task as the eukaryotic genes are not continuous and interrupted by intervening noncoding sequences called ‘introns’.
The ORF present in the isolated sequences was predicted. Total five ORF were identified having sequence length 244, 37, 34, 41 and 34 amino acids Figure 2. The first predicted ORF sequence having amino acid length 244 amino acids is selected based on its length. Based on BLASTP analysis of this predicted sequence was identified as delta 9 acyl-lipid desaturase (Nostoc sp. 36).
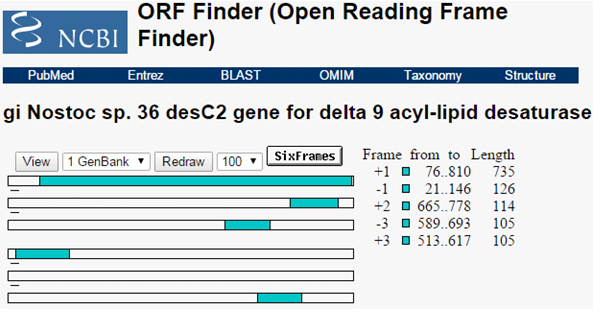
Fig. 2. ORF predicted for the isolated gene sequence
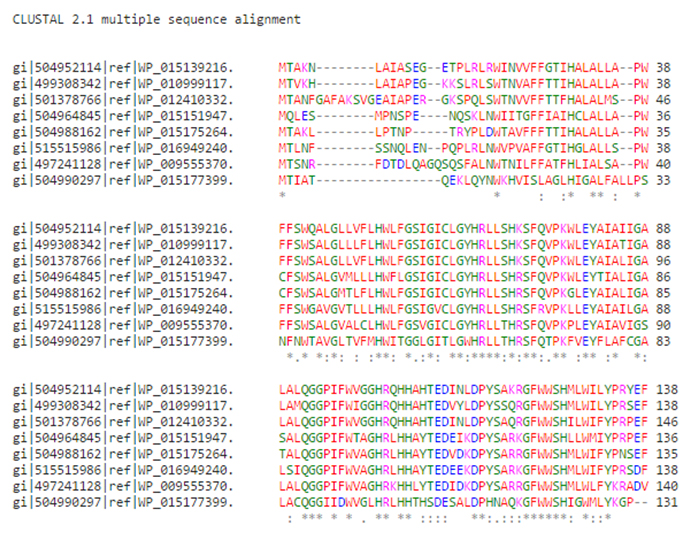
 Fig. 3. Sequence alignment of the selected sequences
Fig. 3. Sequence alignment of the selected sequences
Based on BLASTP analysis of delta 9 acyl-lipid desaturase [Nostoc sp. 36] six similar sequences were identified Table 3.
Table (3):
List of enzymes selected for sequence similarity among species known for lipid production.
S.N0 |
Accession No. |
Enzyme |
Organism |
|---|---|---|---|
1. |
WP_012410332.1 |
delta-9 desaturase |
Nostoc punctiforme |
2. |
WP_015139216.1 |
fatty-acid desaturase |
Nostoc sp. PCC 7524 |
3. |
WP_010999117.1 |
delta-9 desaturase |
Nostoc sp. PCC 7120 |
4. |
WP_016949240.1 |
delta-9 desaturase |
Anabaena sp. PCC 7108 |
5. |
WP_015151947.1 |
fatty-acid desaturase |
Oscillatoria acuminata |
6. |
WP_009555370.1 |
fatty-acid desaturase |
Oscillatoriales cyanobacterium JSC-12 |
7. |
WP_015175264.1 |
Delta-9 acyl-phospholipid desaturase |
Oscillatoria nigro-viridis |
8. |
WP_015177399.1 |
Stearoyl-CoA 9-desaturase |
Oscillatoria nigro-viridis |
When two symbolic representations of DNA or protein sequences are arranged next to one another so that their most similar elements are juxtaposed they are said to be aligned. Multiple Sequence alignment of selected sequences was performed. It was found that sequence similarity was between 46 percent to 85 percent Figure 4.
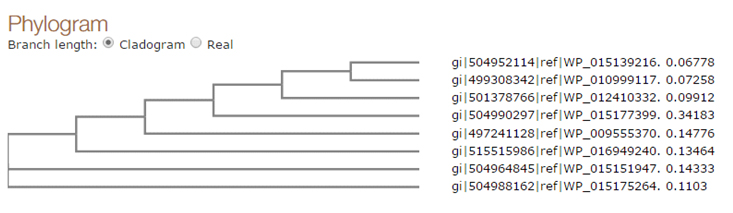
Fig. 4. Phylogenic tree of the selected sequences
Based on phylogenetic analysis it was found that the Fatty-acid desaturase (WP_015139216.1) is most identical with predicted ORF in the sample sequences . The other aligned sequences were clustered in different groups. Fatty-acid desaturase (WP_015139216.1) was clustered with Delta-9 desaturase (WP_010999117.1). Delta-9 desaturase (WP_012410332.1) was found more similar with WP_015139216.1 and WP_010999117.1. Simillarly Stearoyl-CoA 9-desaturase (WP_015177399.1), Fatty-acid desaturase (WP_009555370.1), Delta-9 desaturase WP_016949240.1 and Fatty-acid desaturase (WP_015151947.1) and Delta-9 acyl-phospholipid desaturase (WP_015175264.1) were also found related with the query sequence and clustered in different groups Figure 5. The analysis of fatty acids in different clade should provide useful information to establish the functional role of the enzyme.
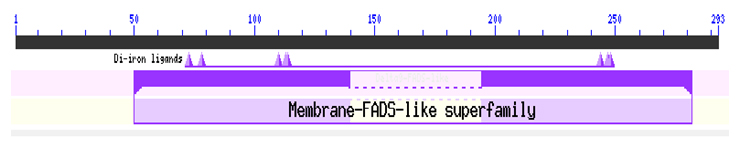
Fig. 5. Predicted domain in the sequence
The putative functional domain was also predicted. It was found that the sequence belongs to the (Membrane_FADS) –like superfamily(Figure 6). Membrane FADSs play an important role in the maintenance of the proper structure and functioning of biological membranes as they play key role in are involved in the initial oxidation of inactivated alkanes (Marchler-Bauer et al., 2011).
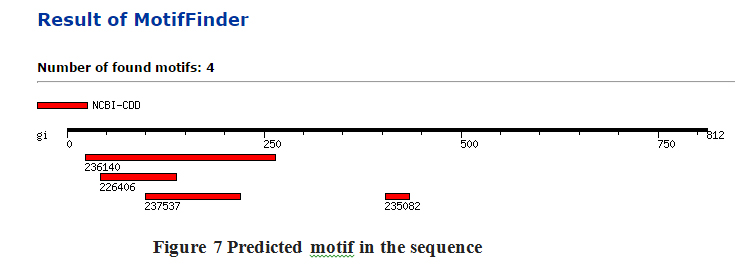
Fig. 6. Predicted motif in the sequence
It was reported that acyl-lipid desaturases contain three histidine clusters, whose structures are unique to individual classes of acyl-lipid desaturases. They are related to the specificity of individual desaturases to the position of carbon atoms in fatty acids at which unsaturated bonds are introduced (Chintalapati et al.,2006).
Motif
Sequence motifs are short, recurring patterns in DNA that are presumed to have a biological function. Often they indicate sequence-specific binding sites for proteins such as nucleases and transcription factors (TF). Others are involved in important processes at the RNA level, including ribosome binding, mRNA processing (splicing, editing, polyadenylation) and transcription termination. Four motifs of different length and positions were identified in the predicted ORF sequence Figure 7. It was found that length of motif were between 31 to 241 base pairs Table 4. All motif are involved in different types of biological activity (Marchler-Bauer et al., 2009).
Table (4):
List of motifs predicted in ORF sequence.
S. No. |
NCBI-CDD I.D. |
Position (base pair) |
Length (base pair) |
Description |
|---|---|---|---|---|
1. |
236140 |
24..265 |
241 |
PRK08026, PRK08026, flagellin; Validated. |
2. |
226406 |
43..139 |
96 |
COG3889, COG3889, Predicted solute binding protein [General function prediction only]. |
3. |
237537 |
99..220 |
121 |
PRK13875, PRK13875, conjugal transfer protein TrbL; Provisional. |
4. |
235082 |
404..435 |
31 |
PRK02888, PRK02888, nitrous-oxide reductase; Validated. |
Metabolic engineering with the gene manipulation technique can be a significant approach to improve the function of enzymes playing key role in fatty acid.
The gene sequence of Acyl lipid desaturases from Nostoc Sp. was analyzed. It’s computational characterization revealed that it belongs to Functional domain (Membrane_FADS) –like superfamily. It was also found that Acyl lipid desaturases is involved in the production of lipids. This study provides information that will help in the regulation of the fatty acid profile and functional characterization of Acyl lipid desaturases involved in the lipid production which can further be used for biodiesel production
ACKNOWLEDGMENTS
The authors are grateful to the Sam Higginbottom Institute of Agriculture, Technology & Sciences, Deemed University, Allahabad for providing the facilities and support to complete the present research work.
- Sing, S. F., Isdepsky, A., Borowitzka, M. A. and Moheimani, N. R. Production of biofuels from microalgae, Mitigation and Adaptation Strategies for Global Change, 2011; 18: 47–72.
- Huang, J. C., Ge, X. J. and Sun, M. Modified CTAB protocol using a silica matrix for isolation of plant genomic DNA. Biotechniques, 2000; 28: 432–4.
- Desjardins, P. and Conklin, D. NanoDrop Microvolume Quantitation of Nucleic Acids J Vis Exp.; 2010; 45: 2565.
- Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975; 94(3):441–448.
- Jin, H. H., Kima, J. H., Sohn, C. H., De Wreede, R. E., Choi, T. J., Towers, G. H. N., Hudson, J. B. and Hong, Y. K. Inhibition of Taq DNA polymerase by seaweed extracts from British Columbia, Canada and Korea. J. Appl. Phycol. 1997; 9: 383–8.
- Hong, Y. K., Sohn, C. H., Lee, K. W. and Kim, H. G. Nucleic acid extraction conditions from seaweed tissues for polymerase chain reaction. J. Mar. Biotechnol. 1997; 5:95–9.
- Potts, M. (1997). ”Etymology of the Genus Name HYPERLINK “http://ijs.sgmjournals.org/content/47/2/584.full.pdf”NostocHYPERLINK “http://ijs.sgmjournals.org/content/47/2/584.full.pdf” (HYPERLINK “http://ijs.sgmjournals.org/content/47/2/584.full.pdf”CyanobacteriaHYPERLINK “http://ijs.sgmjournals.org/content/47/2/584.full.pdf”)”. International Journal of Systematic Bacteriology 47 (2): 584. doi:10.1099/00207713-47-2-584.
- Rasmussen HE, Blobaum KR, Park YK, Ehlers SJ, Lu F, Lee JY. (2008) J HYPERLINK “http://www.ncbi.nlm.nih.gov/pubmed/18287352”NutrHYPERLINK “http://www.ncbi.nlm.nih.gov/pubmed/18287352”. Mar;138(3):476-81
- Marchler-Bauer A et al., ”CDD: a Conserved Domain Database for the functional annotation of proteins.”, Nucleic Acids Res, 2011; 39(D)225-9.
- Marchler-Bauer A et al., ”CDD: specific functional annotation with the Conserved Domain Database.”, Nucleic Acids Res., 2009; 37(D)205-10.
- Marchler-Bauer A, Bryant SH, ”CD-Search: protein domain annotations on the fly.”, Nucleic Acids Res. 2004; 32(W)327-331.
- Aresta M., Dibenedetto A. and Barberio G. Utilization of macro-algae for enhanced CO2 fixation and biofuels production: development of a computing software for an LCA study. Fuel Proc. Technol. 2005; 86: 1679–1693.
- Bligh E. G. and Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959; 37: 911-917.
- Cardozo K. H. M., Guaratini T., Barros M. P., Falcao V. R., Tonon A. B., Lopez N. P., Campos S., Torres M. A., Souza A. O., Colepicolo P., Pinto E. Metabolites from algae with economical impact. Comp. Biochem. Physiol. Comp. Pharmacol. 2007; 146: 66-78.
- Greenwell H. C., Laurens L. M. L, Shields R. J., Lovitt R. W. and Flynn K. J. Placing microalgae on the biofuels priority list: a review of the technological challenges. J. Royal Soc. 2009; doi: 10.1098/ rsif.2009.0322.
- ChintalapatiHYPERLINK “http://www.ncbi.nlm.nih.gov/pubmed/?term=Chintalapati%20S%5BAuthor%5D&cauthor=true&cauthor_uid=16689682” S1, Prakash JS, Gupta P, Ohtani S, Suzuki I, Sakamoto T, Murata N, Shivaji S., A novel Delta9 acyl-lipid desaturase, DesC2, from cyanobacteria acts on fatty acids esterified to the sn-2 position of glycerolipids, BiochemHYPERLINK “http://www.ncbi.nlm.nih.gov/pubmed/16689682” J. 2006 Sep 1;398(2):207-14.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


