ISSN: 0973-7510
E-ISSN: 2581-690X
Detection of phytopathogens that are involved in causing disease symptoms in plants and crops is of prime importance as a key step in disease treatment and management. Ziziphus mucronata is a species endemic in temperate and tropical climates and used traditionally in the treatment of infectious diseases. The common bean (Phaseolus vulgaris) is a rich source of nutrients for the human diet. Just like most crops, it is not immune to fungal diseases and reports had been received of P. vulgaris showing signs on disease. The aim of this study was to isolate and characterize the fungal species associated with the disease symptoms in the Z. mucronata and P. vulgaris. Fungal species where isolated from surface-sterilised symptomatic bark of Z. muconata and fresh green bean pods. These where grown on petri dishes containing Potato Dextrose Agar and incubated at room temperature. Pure cultures where then obtained by transferring small segments of fungal growth to a new petri dish that contained PDA. During DNA extraction the pure cultures where first homogenized using liquid nitrogen and then the rest of the extraction carried out as stipulated in the Zymo extraction kit. The Nanodrop was used for quantifying the DNA and amplification of the conserved Internal Transcribed Spacer (ITS) region of Ribosomal RNA genes was carried out using ITS1 and ITS2. The PCR products were sequenced at Inqaba Biotech Industries in South Africa. The obtained sequences were then compared by alignment with known sequences in the Genbank using Basic Local Alignment Search Tool (BLAST). The BLAST searches were able to reveal the fungi isolated from the Z. mucronata as Fusarium penzigii and Fusarium dimerum while the fungi isolated from P. vulgaris shown to be Phoma destructiva, with 100 %, 95% and 100% sequence similarity respectively. The next step in this work is carry out Koch‘s postulates to determine which of this fungi is the causal agent of the observed diseases symptoms in order to start a targeted diseases management programme.
Ziziphus mucronata, Phaseolus vulgaris, Internal Transcribed Sequence, Alignment, BLAST, Fusarium dimerum, Fusarium penzigii, Phoma destructiva
The Ziziphus mucronata also known as the Buffalo thorn is a small tree with a spreading canopy that has characteristic zigzag young twigs with two significant thorns at the node, with one facing forward and the other backwards. Z. mucronata is a multipurpose tree with a variety of traditional importance such as being believed to protect from lighting in Botswana (Mazibuko, 2007). (Mazibuko, 2007) stated that many parts of the plant are used in ethno medicine with the tannins playing a role in dysentery treatment and the roots being used in East Africa to treat snake bites, also went on to say that these properties have been attributed to the peptide alkaloids and antifungal properties isolated from the bark and leaves.
The common bean (Phaseolus vulgaris) also called the green bean is a herbaceous legume that has its origin in South and Central America, it is now cultivated worldwide for its high nutritious value (Earth, 2011). The activity of bacteria found in its root nodules is able to confer nitrogen fixing properties to the bean. Due to poor management, the beans may be exposed to high moisture and temperature during storage and harvest, this may provide a suitable environment for fungal growth which might lead to contamination by mycotoxins (Scussel, 2000).
This research aimed to address the problem of lack of information on identity of plant diseases in Z. mucronata and common green beans. Fungi cause a wide range of crop and plant diseases worldwide. The Ziziphus mucronata is one of the important ethno medical plants in Africa and Asia. It has medical properties such as antitumor, anticancer and anti-diabetic. There have been various reports on green beans showing signs of diseases and a few studies have been conducted to follow up these reports hence the reason behind this study focusing on fungi causing diseases on the common green beans.
Sampling Area
Symptomatic plant tissue from Z mucronata was collected from the University of Namibia while the diseased green beans samples where obtained in local green grocer shops. To minimize destruction of the plant, nondestructive sampling was carried out.
Obtaining and Sterilization of Samples
Branches that showed disease symptoms where obtained from the tree and fresh green beans pods where also obtained. The symptomatic tissues where then surface sterilized according to the procedure used in (Garas, et al., 2012).
Isolation of Fungi
After the sterilization, a piece of crop tissue from the bean pod and from the Ziziphus mucronata stem with fungal growth on it was cut and transferred onto an agar plate containing Potato Dextrose Agar. The fungi was then incubated at room temperature of 25oC and left to grow for 5 days. Sub culturing was performed where required and after pure cultures of the fungi where obtained from single spores, these cultures had to be stored in 30% glycerol.
DNA Extraction and PCR Amplification
DNA was then extracted from the pure cultures using the ZYMO Fungal DNA extraction kit according to the manufacturer’s manual. Gel electrophoresis was carried out so as to confirm the presence or absence of fungal DNA which was evident from the bands. Further quantification was then carried out using a Nano drop. PCR was carried out with the following PCR mixtures: Buffer (12.5µl), DNA (4µl), sterile water (5.5µl) and primers (3µl) giving a total master mix mixture of 25µl. The Amplification was done using ITS1 and ITS2 primers at a primer concentration of 0.8uM with the following profile: Pre-denaturation of DNA, 94°C for 4 minutes, Denaturation of target DNA; 94ºC for 20 seconds,
Primer annealing; 59ºC for 30 seconds, Primer elongation; 72ºC for 1 minute, Final elongation; 72ºC for 10 minute and Final hold at 4ºC The amplicons where then sent to Inqaba Biotech Industries for automated sequencing of the ITS region.
DNA Sequence Analysis
ITS 1 and ITS 2 primers that had been used for amplification were used for sequencing in both forward and reverse directions with the obtained sequences being used to perform Blast searches in the NCBI Genbank. The integrity of the sequences was ensured by eye inspection.
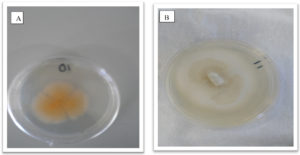
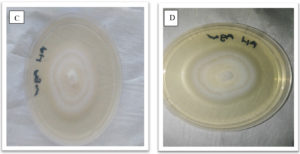
Fungal cultures where obtained from tissue of the Z. mucronata showing symptoms of infection and sub culturing was carried out until pure cultures were obtained as depicted in figures 2 and 3. Fusarium dimerum could be identified from the plates due to the morphological orange color it grew with.
Fig. 1. Showing a Ziziphus mucronata tree at the University of Namibia showing symptoms of disease by the yellowing and loss of leaves
Fig. 2. Showing morphological appearance of isolates from Z. mucronata. (A- Fusarium dimerum and B-Fusarium penzigii)
Fig. 3. Showing morphological appearance of isolates from P. vulgaris. With both C and D having come from Phoma destructiva
DNA was then extracted from the pure cultures and Gel electrophoresis was run and a Thermo Scientific NANADROP 2000c a product of MET supplied by Bio Dynamics used to quantify the DNA.
The obtained amplicons were then cleansed and sequenced. After BLASTN the fungi responsible for the disease symptoms in the Z. mucronata where revealed by 100% and 94% similarity as Fusarium penzigii and Fusarium dimerum respectively. While Phoma destructiva was revealed to be the isolate from P. vulgaris.(Maier, et al., 2014) who carried out one of the few available studies on diseases of the Z. mucronata concluded that Coniodictyum chevalieri was responsible for the diseases symptoms that where observed in the Z. mucronata found in the Kruger National Park in South Africa. The authors also went on to state that C. chevalieri has a unique biology, regarding its biogeography, ecology and host specificity, as it only infects Z mucronata.
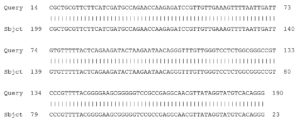
Fig. 4. Partial display of local pairwise sequence alignment of ITS regions showing 100% similarity to Fusarium penzigii (Query) sequence that was against the subject sequence in the GenBank database
Fig. 5. Partial display of local pairwise sequence alignment of ITS regions showing 95% similarity to Fusarium dimerum (Query) sequence that was against the subject sequence in the GenBank database and 5% mismatch
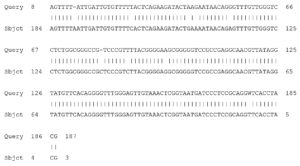
Fig. 6. Partial display of local pairwise sequence alignment of ITS regions showing 100% similarity to Phoma destructiva (Query) sequence that was against the subject sequence in the GenBank database
Most studies that have been carried out on the Z. mucronata have been focused on mostly its ethno medical importance and a few studies have been documented on the diseases that affect the plant. Fusaruim species have proved to the causal agents of most plant and crop diseases not neglecting their effects on certain opportunistic infections in humans as indicated in (Salah, et al., 2015) where they are indicated to be found in wounds, blood, nails, cornea, urine and skin.
Fusarium dimerum had been believed to be decaying plant saprothrophs, take (Gerlach & Nirenberg, 1982) who were unaware of F. dimerum causing any plant diseases when they carried out their studies. The findings of this study thus refutes that belief as it has shown that the fungi is also associated with disease symptoms in plants. It is also now known as a species complex normally abbreviated (FDSC) which contains at least 12 lineages, of which F. dimerum sensu stricto, F. delphinoides, and F. penzigii are common distinct species according to (van Diepeningen, et al., 2014), (Josef, et al., 2008) also goes on to support this claim by stating that Fusarium dimerum is known to comprise of at list 12 phylogenetically distinct species and is only known for its anamorph.
Fusarium penzigii was named in honor of an Italian mycologist by the name of Albertus Giulio Ottone Penzig. Just like the F. dimerum species complex it is widely known to be a soil and dead plant substrata pathogen or as an agent of trauma-related eye infections of humans so this report is one of the rare ones that indicates its role in causing plant diseases.
The results showed that Phoma destructiva was responsible for the disease symptoms on the green beans. This observation is one that has not been widely documented in literature as P. destructiva has mostly been reported to cause disease symptoms in tomatoes. Phomopsis blight of tomatoes is known to be caused by the fungi Phoma destructiva, it causes rot spots that are usually sunken, light colored at margins and leathery, the fungus also produced pcynidia in dark portions of the tomato (Wani, 2011).
This study was limited in that there was not enough time available to collect the seedlings of the Ziziphus mucronata and carry out Koch’s postulates so as to test the obtained species for pathogenesis. The results of this study support the hypothesis that fungi was responsible for the symptoms observed on the Ziziphus mucronata and the Phaseolus vulgaris. With the fungi suspected to be responsible being isolated and characterized to the species level which was the main objective of this study. It can thus be concluded that Z. mucronata is susceptible to infection by Fusarum penzigii and Fusarium dimerum while the common green beans is susceptible to Phoma destructiva infection. To be able to confidently point out the isolated fungi as the casual agents, a couple of more experiments should be designed and further extensive studies should be carried on the isolates.
The results of this study supported the hypothesis that fungi was responsible for the symptoms observed on the Ziziphus mucronata and the Phaseolus vulgaris. With the fungi suspected to be responsible being isolated and characterized to the species level which was the main objective of this study. It could thus be concluded that Z. mucronata was susceptible to infection by Fusarum penzigii and Fusarium dimerum while the common green beans was susceptible to Phoma destructiva infection.
- Avise, J. C., Twenty-five key evolutionary insights from the phylogeographic revolution in population genetics. SpringerLink, 2007; pp. 7-21.
- Benny, G. L., Humber, R. L. & Morton, J., n.d. Zygomycota;Zygomycetes. SpringerLink, pp. 113-146.
- Brandt, M. E., Lockhart, R. S. & Warnock, D., 2010. Laboratory Aspects of Medical Mycology. Atlanta: SpringerLink.
- Capote, N., Pastrana, M. A., Aguado, A. & Torres, S. P., 2012. Molecular Tools for identification of plant pathogenic fungi and fungicide resistance. SpringerLnk.
- Chi, M.-H., Park, S.-K. & Lee, Y.-H., A Quick and Safe Method for Fungal DNA Extraction. Plant Pathology, 2009; 25(1), pp. 108-111.
- Christou, P. & Twyman, R., The Potential of genetically enhanced plants to address food insecurity. NCBI, 2004; pp. 23-40.
- Crous, P. et al., How Many Species Of fungi Are There At the Tip Of Africa. NCBI, 2006; pp. 13-20.
- Earth, A., Phaseolus vulgaris, s.l.: The Plant Encyclopedia 2011.
- Garas, L. S., Uzabakiriho, J. D. & Chiwmamurombe, P. M., Isolation and identification of Fungal Species Associated with Gall Formation on Acacia mellifera in the Western Windhoek. Journal of pure and applied microbiology, 2012; pp. 713-716.
- Gerlach, W. & Nirenberg, H. I., The genus Fusarium—a pictorial atlas. Mitt Biol Bundesanst Land-Forstw Berlin-Dahlem, 1982; 209, pp. 1-406.
- Goud, J. C. & Termorshuizen, A. J., Quality of methods to quantify microsclerotia of Verticulum dahliae in soil. European journal of plant pathology, 2003; pp. 523-534.
- Guarro, J., Gena, J. & Stchigel, A., Developments in Fungal Taxonomy. American Society for Microbilogy, 2006; pp. 457-484.
- Hoog, S. & Guarro, J., Atlas of Clinical Fungi. cabdirect 2000.
- Josef, H. et al., Taxonomy and phylogeny of the Fusarium dimerum species group. Mycologia, 2008; 101(1), pp. 44-70.
- Kang, S. et al., The promise and pitfalls of sequence-based identification of plant-pathogenic fungi and oomycetes. Phytopathology, 2010; 100(8), pp. 732-737.
- Kurtzman, C. P. & Robnet, C. J., Phylogenetic relationships among species of Saccharomyces, Schizosaccharomyces, Debaryomyces and Schwanniomyces determined from partial ribosomal RNA sequences. 2004; Wiley.
- Maier, W. et al., A disease epidemic on Zizyphus mucronata in the Kruger National Park caused by Coniodictyum chevalieri. Studies in Mycology, 2014; 55, pp. 279-288.
- Mazibuko, N., Ziziphus mucronata Willd. South African Biodiversity Institute 2007.
- Mullis , K. B. & Faloona, F. A., Specific Synthesis of DNA in vitro via a polymerase-catalyzed chain-reaction. Methods in Enzymology, 1987; 155, pp. 355-350.
- Narayanamasamy, P., 2011. Microbial Plant Pathogens-Detection and Disease Diagnosis. s.l.:s.n.
- Oerke, E. C., 1994. Crop Protrction and Crop Production. cabdirect.
- Olaguyigbe, O. O. & Afolayan , A. J., Antimicrobial Potency of the ethanolic crude bark extract of Ziziphus mucronata Willd. Academic Journals, 2012; 6(10), pp. 724-730.
- Salah, H. et al., Phylogenetic Diversity Of Human pathogenic Fusarium and emergence of uncommon virulent species. Journal Of Infection, 2015; 71(6), pp. 658-666.
- Scussel, V. M., Micotoxinas em Micotoxinas e Armazenagem de Graos. VMS, 2000; p. 382.
- Soledad, M. et al., The Phytopathogenic fungus Alternaria brassicicola: Phytotoxin production and phytoalexin elicitation. elsevier 2009.
- Tharreau, D., Silue, J. & Notteghem, L., n.d. Genetic Study of Host-Parasite Relationship in the Oryza Sativa-Magnaporthe Grisea Pathosystem. SpringerLink, pp. 49-49.
- van Diepeningen, A. D., AL-Hatmi, A. M., Brankovics, B. & de Hoog, G. S., Taxonomy and Clinical Spectra of Fusarium Species: Where Do We Stand in 2014?. SpringerLink, 2014; 1(1), pp. 10-18.
- Wani , A. H., An overview of the fungal rot of tomato. Mycopath, 2011; 9(1), pp. 33-38.
- Zhang, S. et al., Floral transition in maize infected with Sporisorium reilianum disrupts compatibility with this biotrophic fungal pathogen. SpringerLink, 2013; pp. 1251-1266.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.