ISSN: 0973-7510
E-ISSN: 2581-690X
This study aims to isolate and characterise the Pink Pigmented Facultative Methylotrophs (PPFM) bacteria from the leaves of common Malaysian table salad (ulam). The colonies of PPFM bacteria were obtained using selectively modified Pseudomonas agar based on the appearance of the pink pigment colony. The three selected isolates labelled OJ4, OJ154 [Oenanthe javanica (Blume) DC.] and ML8 [from Melicope lunu-ankenda (Gaertn.) T. G. Hartley] were chosen for characterisation. The result showed the PPFM bacteria colonies (log CFU/g) at the leaf surface of C. caudatus are 4.4 ± 0.1, significantly higher than O. javanica, 3.8 ± 0.2, and M. lunu-ankenda, 3.2 ± 0.1. The selected isolates belong to the Gram-negative group, motile with rod shape with size [length (l) × width (w)] in µm unit 4.3 ± 1.1 × 1.6 ± 0.6, 5.4 ± 0.2 × 1.2 ± 0.1, and 3.5 ± 0.7 × 1.0 ± 0.0, respectively. They show positive urease, catalase, and oxidase activities, while none of them can degrade starch, gelatine, or cellulose, as well as glucose fermentation (MR test) and metabolism actions (VP test), producing indole and hydrogen sulphide gaseous. Only isolate OJ154 demonstrates positive casein hydrolysis and nitrate reduction activities, while only isolate ML8 can utilise citrate but not in lipid degradation. Their sequence analysis of the 16S rRNA indicated that OJ4 and ML8 are Methylobacterium radiotolerans with a similarity of 99%, whereas, OJ154 is Methylorubrum salsuginis with a similarity of 99%. To conclude, PPFM bacteria from the leaves of C. caudatus, O. javanica, and M. lunu-ankenda have been isolated and characterised in particular.
Ulam, Pink Pigmented Facultative Methylotrophs, Bacteria Isolation, Bacteria Characterisation, Biochemical Assay, Molecular Identification
There are numerous vegetable kinds available on the market that can be consumed in cooked form, like broccoli, or in raw form, such as salad. Their nutritional value may be variable depending on how they are prepared to eat. Previous studies have shown that some vegetables reduce their phytochemical contents, for example, polyphenols, carotenoids, and ascorbic acid, after cooking compared to raw.1 Another study revealed that raw vegetables and salad cause an increase in serum levels of vitamins, including vitamin C, vitamin E, folic acid, β-carotene, and lycopene, in their consumers’ bodies.2
Hence, consuming fresh and raw vegetables like ulam can be one of the best choices for a healthy diet, as its nutritional value, including phytochemical contents remains unchanged due to no heat applied. Malaysian table salad also known as ulam, is commonly eaten freshly (raw) with rice as a side dish.3 Throughout generations, ulam has been consumed by mankind due to its believed possession of various therapeutic benefits, including the prevention of degenerative diseases, the delay of ageing, and the improvement of overall health.2,4
Ulam may consist of young leaves, shoots, flowers, fruits, roots, and rhizomes as part of vegetables.5 Several examples of common ulam consumed by Malaysians are pegaga (Centella asiatica), petai (Parkia speciosa), ulam raja (C. caudatus), and selom (O. javanica).6 Fascinatingly, much scientific research has shown the benefits of the phytochemicals contained in ulam to its consumers. For example, C. caudatus, O. javanica, and M. lunu-ankenda have nutritional values like carbohydrates, proteins, minerals, and vitamins.4,7,8 Their structure also contains phytochemicals such as phenolic compounds of caffeoylquinic acids and proanthocyanidins (C. caudatus); flavonoids, phenolic, and volatile oils that contribute to its aromatic smell (O. javanica); and isorhamnetin, skimmianine, and leptonol (M. lunu-ankenda).4,8-10 The chemical compound content of the 3 types of ulam are responsible for giving similar benefits to health, such as anti-inflammatory and antioxidant effects.4,8,10
Besides, the raw condition of vegetables also preserves symbiotic microorganisms that are present in their structure. These symbiotic microorganisms may add value to the quality of the raw vegetables themselves. Typically, the inhabitants of symbiotic microorganisms who occupy the leaf surface of ulam are identified as PPFM bacteria. It is the methylotrophs that inhabit the phyllosphere, mostly on the leaf surfaces of plants, including ulam leaves. The group is named methylotrophs and is known as PPFM bacteria, classified into the genus Methylobacterium.11,12
Uniquely, Pink Pigmented Facultative Methylotrophs (PPFM) bacteria could use single-carbon compounds as well as multi-carbon growth substrates for their energy and carbon source solely,13 allowing them to consume ‘food’ such as methanol released by plants and, in return, providing essential compounds for the plant’s development.14 A valuable compound identified as pyrroloquinoline quinone (PQQ) was formed by them during the oxidation of the single compound. This compound also has anti-diabetic and anti-oxidant activities. Additionally, this kind of symbiotic bacteria does no harm to their host plants, making their bioactive compounds likely to be less toxic to humans, which is important for application.15,16
Previous studies have stated the same or similarity in a few symbiotic microbes’ cell structures with their host plants.17,18 For example, compounds, namely camptothecin and its structural analogues produced by endophytic fungi are also synthesised by their host.19-22 Moreover, Sarjono et al.23 reported that both the extracted isolated bacteria’s compound and their host plant’s leaf (Carica papaya L.) contain alkaloids, flavonoids, and tannins. Hence, the knowledge discovered from this study regarding the properties of PPFM bacteria obtained from ulam may improve the quality of food in general. In other words, people who consume ulam may get both benefits from the phytochemicals of ulam and bioactive compounds from the PPFM bacteria residing in its structure.
As mentioned previously, these bacteria are a type of microorganism that can utilise one-carbon compounds as a sole source of carbon and energy, as well as other multicarbon substrates present ubiquitously in nature, such as plant leaves, in symbiotic relationships.24 To date, numerous studies have been conducted to identify the benefits of the pigments extracted from this bacterial species, but none of them focused on the same bacteria that originated from Malaysian ulam leaf as well as its potential application as a food colourant. Hence, this study emphasised the PPFM bacteria properties (number of colonies, biochemical and molecular characterisation) together with their extracted pigments for bioactivity (antioxidant) performance evaluation from selected ulam leaf species for further study.
Previous studies have shown bacteria cells or their extracted compounds have been implemented as food, such as a supplement, not only for animals but also for humans. For example, Jones et al.25 reported that several microorganism species, including PPFM, have been genetically modified to produce carotenoids and taurine or its precursors in their aquaculture stock.26 Moreover, the biomass of PPFM, specifically from M. extorquens, has been applied in trout feeds as an alternative protein source, replacing soybean meal.27
In addition, one example of a bacteria compound (PQQ) has been extracted from Hyphomicrobium denitrificans CK-275 fermentation with the trade name BioPQQTM and is considered safe to consume at the intended conditions approved by the European Food Safety Authority (EFSA).28 However, the further objective of this study concerns the application of pigment extracted from PPFM to food. Therefore, the pigment’s toxicity level has been tested and can be considered safe for consumption under certain concentrations (manuscript publication in process).
Materials
The 3 samples of fresh ulam leaves, namely C. caudatus, O. javanica, and Melicope lunu-ankenda (Gaertn.) T. G. Hartley leaves, were obtained from both wet and night markets (each approximately 150-200 g per species) at Sri Serdang, Selangor, Malaysia. For chemicals, phosphate buffer solution (PBS) and sulfanilic acid were obtained from Sigma (Missouri, United States), while Pseudomonas agar F, peptone was bought from Difco™, BD (New Jersey, US). Agar-agar types I, nutrient gelatin, Simmons citrate agar (SCA), sulphide indole motility (SIM) medium, and nitrate broth were purchased from HiMedia (Mumbai, India). Glycerol (anhydrous), skim milk powder, potassium chloride, sodium nitrate, carboxymethyl cellulose (CMC), N-cetyl-N,N,N,trimethyl ammonium bromide (CTAB), potassium dihydrogen phosphate, phenol red, methylene blue, 1-naphthol, methyl red, Kovac’s (indole reagent) solution, D(+)-glucose (anhydrous), potassium sulphate, and 1-naphthylamine were purchased from R & M Chemicals (Essex, UK). N,N-Dimethyl-1,4-phenylenediamine oxalate (compound) was obtained from Sigma-Aldrich (Missouri, United States).
Furthermore, crystal violet, safranin O, and zinc powder were brought from Bendosen (Johor, Malaysia), while ethanol (95%), magnesium sulphate, di-potassium hydrogen phosphate (phosphate buffer), and glacial acetic acid were obtained from Systerm (Selangor, Malaysia). Ammonium oxalate, soluble starch, urea, potassium hydroxide, and hydrogen peroxide were bought from Chemiz (Selangor, Malaysia). Sodium chloride and iodine pearl were purchased from QRëC™ (Bangkok, Thailand). Nutrient agar, tributyrin agar, yeast extract, and potassium iodide were purchased from Fisher Scientific (New Hampshire, US). In addition, trypticase soy agar (TSA) and disodium phosphate were bought from Merck (Darmstadt, Germany), cellulose nitrate membrane filter was purchased from LabServ® (Massachusetts, United States), and BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) was from Thermo Fisher Scientific, Massachusetts, United States. For the experiment, all chemical reagents were carefully selected to be of analytical grade, and all aqueous solutions required for the study were meticulously prepared using distilled water.
Methods
Isolation of PPFM bacteria
Firstly, according to Madhaiyan29, Mizuno et al.30, Sanjee et al.31, and Kaparullina et al.,32 an amount of 1 g of the fresh ulam leaves were weighed, washed with sterilised distilled water, and mashed using a mortar and pestle, serially diluted, and 100 µl of the aliquot from each dilution were transferred on the sterile modified Pseudomonas agar (MPA) medium using the spread plate method.33 The preparation of 1 L of MPA is done by mixing 3.8 g of Pseudomonas agar, glycerol 1.5 ml, and agar Type I 18.9 g.
Pseudomonas agar F was used in this study rather than others because Methylobacterium species have sufficient nutrients to grow on the media. Its basic composition, which contains glycerol, proteose peptone no. 3, pancreatic digest of casein, dipotassium hydrogen phosphate, and magnesium sulphate heptahydrate, resembles King’s B agar medium composition except for the addition of pancreatic digest of casein. Both of these media were used to isolate, cultivate, and differentiate Pseudomonas species generally and have been applied to PPFM species since these bacteria have similar characteristics to Pseudomonas species.34,35 Moreover, the pigmentation of PPFM bacteria on Pseudomonas agar is deep, similar to when growing on glycerol-peptone agar, making it suitable for subculture but not for culture enrichment.36
Next, the plates were incubated in aerobic conditions at 30°C for 3-7 days. This assay was performed 3 times with duplicates (n = 3×2). The presence of pink-coloured colonies that appear on the MPA medium was counted for CFU value. Afterwards, the streak plate method was employed to isolate and purify a total of 100 pink-coloured bacterial colonies from each type of ulam, with each colony displaying distinct characteristics. Once purified, the colonies were carefully stored at a temperature of 4°C for future use.33
Morphological and biochemical characterisation
Among the 100 PPFM colonies that have been isolated and purified randomly from each type of ulam leaf (ulam raja, selom, and tenggek burung), 40 pure cultures from them (each ulam type) were selected based on their colour intensity (from pale pink to deep pink) and colony size differences (big and small). Then, they proceeded with further examination steps involving microscopic observation as well as several biochemical assays, and those experiments were done in triplicate. All the procedures described for morphology and biochemical activity analysis except lipolytic and cellulose activities were referred to by Barrow and Feltham37 in general.
Gram staining
To differentiate the main class of bacteria, the Gram staining method with a slight modification from Engelkirk et al.38 and the U.S. Food and Drug Administration39 was used. The slide containing Gram-stained bacteria cells was examined under a compound microscope for Gram identification. The bacteria cells that retained the crystal violet-iodine complex during the decolorization step are considered as Gram-positive and vice-versa. The Gram-negative bacteria cell was stained pink to red due to the safranin (a red dye).38
Motility and size observation
Initially, by referring to Laxmi et al.40 and Pattanashetti,41 a loopful of isolated bacteria was placed on a clean glass slide, and a small drop of distilled water was added. The mixture was then evenly smeared and covered with a glass cover slip. After that, the prepared slide was labelled, observed, and measured microscopically, including body size, and the data was recorded.
Cell shape examination
For cell shape examination based on Pattanashetti,41 the same isolate and steps from the previous experiment (motility) were used except for the application of a methylene blue solution for staining purposes. The staining procedure begins with placing a drop of methylene blue solution on a glass slide containing a thin film of isolate and leaving it for a few seconds. Then, the slide was covered with a glass cover slip, and the shape was examined. Finally, the data was recorded.
Starch hydrolysis
Starch agar medium was prepared according to Madhaiyan.29 The assay started with streaking a linear line of isolate culture on the medium, followed by incubation at 35°C for 48 hours. Then, the medium was flooded with Gram’s iodine, and the reaction was observed. The presence of a clear zone surrounding the colonies indicates the hydrolysation of starch and is considered a positive reaction.
Casein hydrolysis
The agar preparation method was modified from Atlas42 and Madhaiyan.29 The isolate culture was streaked on the prepared medium and incubated at 35°C for 48 hours. Clear zone formation around the colonies gives positive results for casein hydrolysis.
Gelatin hydrolysis
The gelatin medium was ready, as stated by Madhaiyan29 and the isolate culture was stabbed deep into it. The medium was then incubated at 35°C for 48 hours. After that, the medium was put back into the chiller for 48 hours to allow the gelatin to solidify for a second time. Then, the result was observed directly. In detail, a gelatin medium with no solidification is a positive result as the process of gelatin liquefaction activity occurs.
Cellulose degradation
According to Aneja,43 the Czapex mineral salt agar medium was prepared to perform the cellulose degradation test. The isolate culture was streaked on the plates and incubated at 35°C for 48 hours. As a result, the presence of clear zones surrounding the colonies after flooding with 1% of CTAB solution onto the agar surface showed a positive reaction.
Citrate utilisation test
To observe citrate utilisation, SCA was prepared based on Madhaiyan.29 The isolate culture was inoculated into the medium and incubated for 48 hours at 35°C. The positive result was illustrated by the changing colour of the medium, from green to blue.
Lipolytic activity
The ready-made tributyrin agar medium that was used for lipolytic activity assessment, as referred to by Madhaiyan.29 The isolate culture was streaked on the medium and incubated at 37°C for 48 hours. The formation of a clear zone surrounding the colonies is taken as a positive reaction.
Urease test
Based on Atlas,41 the urea broth medium was prepared as described. The isolate culture was inoculated in the broth medium and incubated for 24 hours at 35°C. Changing the colour of the urea broth medium from orange to pink/red colour indicated a positive reaction.29
Methyl red (MR) test
The broth medium for the MR test was also used for the Voges-Proskauer (VP) test. Therefore, the name is Methyl Red-Voges-Proskauer (MRVP) medium. The MRVP medium was prepared according to Atlas41 with a slight modification. In a separate process, the MR reagent was prepared, following the guidelines outlined by FDA.39 Following a procedure similar to the previous step, the isolated culture was inoculated into the MRVP broth medium and allowed to incubate for 48 hours at a temperature of 35°C. The result was determined by adding a few drops of the MR reagent into the broth, and the presence of a red colour indicated a positive reaction.29
Voges-Proskauer (VP) test
As per the guidelines provided by FDA,39 the VP reagents were prepared. Utilising the same broth culture prepared for the MR test as described earlier in the section Methyl Red (MR) Test, 0.6 ml of the first reagent was added to 1 ml of the broth culture, followed by the addition of 0.2 ml of the second reagent. The broth culture was shaken during each reagent addition. The reaction was observed after 5 minutes, and the presence of a red colour indicated a positive reaction.44
Nitrate reduction test
The preparation of the nitrate reduction test in this study was based on Faddin’s45 method. As for the reagents, a 0.5% solution of 1-naphthylamine (reagent A) was prepared together with reagent B (0.8% sulfanilic acid). In the test, the isolate culture was inoculated into the nitrate broth and incubated at 35°C for 24 hours. The nitrate reduction reaction of the culture was determined by adding an equal amount of both reagents to the broth and gently shaking to mix them. After 1 to 2 minutes, the appearance of a red colour indicated a positive reaction. If no colour changes were observed (a negative reaction), a further technique was employed by adding a small amount (approximately 20 mg) of zinc dust to the broth, and the colour development was observed after 5 to 10 minutes. The presence of a red colour indicated a negative reaction.
Catalase activity
A modified method based on the Madhaiyan29 protocol was used, where a small number of isolate cultures from an MPA plate were transferred onto a glass slide using a sterile cotton bud. This was followed by the addition of 3% hydrogen peroxide (a drop) to the cultures. The presence of effervescence (bubble formation) indicates a positive reaction.
Hydrogen sulphide production
By referring to Madhaiyan2,9 SIM agar medium was used for the hydrogen sulphide production test. The isolate culture was carefully stab-inoculated into the medium, ensuring proper placement along the line, and incubated for 48 hours at 35°C. Subsequently, the formation of black colouration along the stab inoculation line was observed. The positive reaction was identified by the presence of the black line.
Indole production
Differently, the medium used for indole production is not glucose tryptone broth, as done by Madhaiyan,29 but using the same medium in a hydrogen sulphide production test based on Faddin.45 Then, the result was examined by placing 0.3 ml of Kovacs reagent into the medium, and the change in colour was documented. The reddening of the Kovacs reagent’s layer within a few minutes was considered a positive reaction.
Oxidase test
To examine the oxidation reaction, a TSA medium was prepared for isolate culture growth.29 Separately, reagents named Gaby and Hadley were prepared for the test, according to Faddin.45 In the experimental procedure, the isolate culture was streaked onto a plate and incubated at 37°C for 48 hours. Following incubation, the oxidation reaction was tested using a direct plate procedure. This involved adding 2 to 3 drops of the reagents directly onto the colonies and observing any colour changes within 10 seconds, as per Public Health England46 guidelines. The appearance of a blue colour shows a positive reaction.47
Molecular characterisation
For these assays, only 3 selected pure cultures of PPFM bacteria colonies from the isolate collection (OJ4, OJ154, and ML8) have proceeded for further examination steps and discussion. Those 3 isolates mentioned were selected due to their highest cell number production at 3 days of growth culture (preliminary study) instead of their pigment colour intensity. The assays involved are 16S rRNA gene amplification and DNA sequencing analysis based on Reysenbach et al.48 The full-length 1.5 kb 16S rDNA of the PPFM bacteria was amplified by using universal primers 27F and 1492R. The amplification reaction was performed in a total volume of 25 μl, which included gDNA purified using an in-house extraction method, 0.3 pmol of each primer, deoxynucleotides triphosphates (dNTPs) at a concentration of 400 μM each, 0.5 units (U) of DNA polymerase, supplied PCR buffer, and water.
The PCR amplification procedure was directed as follows: an initial denaturation step at 94°C for 2 minutes was followed by 25 cycles of annealing and extension of the amplified DNA. Each cycle consisted of denaturation at 98°C for 10 seconds, annealing at 53°C for 30 seconds, and extension at 68°C for 1 minute. Subsequently, the PCR products were purified using a standard method and subjected to direct sequencing with primers 785F and 907R, employing the BigDye® Terminator v3.1 Cycle Sequencing Kit from Applied Biosystems. The resulting 16S rRNA gene sequences were assembled using SnapGene® software and analysed using the Basic Local Alignment Search Tool (BLAST) against PPFM bacteria strains available in the National Centre for Biotechnology Information (NCBI) database library to identify the specific gene sequence. The individual gene sequences for all strains were aligned using MUSCLE, and then the neighbor-joining (NJ) phylogenetic tree was generated using Mega 10.2.6 software.49
NJ can be defined as two taxa that are neighbors in a tree if the path between them contains only one node.50 The algorithms for the construction of the phylogenetic tree in this study use NJ due to its characteristics: few assumptions, fast operation, and high accuracy, based on the distance between taxa. They can be used with a variety of models.51,52 This method usually constructs different phylogenetic trees for the same dataset with differences in input order, also known as ‘tied trees’.52 In other words, it provides an unrooted tree and a single resultant tree.51
In detail, the individual taxa are iteratively grouped together, forming larger and larger clusters of taxa. Furthermore, the NJ does not assume a molecular clock, but it assumes that observed distances are near an additive metric, differently from the Unweighted Pair Group Method with Arithmetic Mean (UPGMA). Given an additive metric, the NJ method identifies the correct tree, and it also correctly reconstructs trees if additivity only holds approximately. As neighbor relationships of nodes in a binary uniquely define the tree topology, successively identifying neighbors is a way to reconstruct the tree. In each iteration of the NJ algorithm, every pair of taxa is evaluated for being neighbors, and if so, they are grouped together to form a new taxon for the next iteration.50
Based on all the described methods, the experimental procedures to isolate and characterise PPFM bacteria from ulam leaf in this study involve isolation and purification using semi-selective media, characterization through physical and biochemical tests, followed by partial 16S rRNA gene sequence analysis of randomly picked up pink pigmented colonies (the first isolation step) and only the data of the 3 isolates was discussed.
The PPFM bacteria from ulam leaves were isolated, followed by their mean CFU per gram calculation. Several physical characteristics of the selected PPFM bacteria were identified through their Gram staining properties, motility, cell size, and shape. Also, its 14 biochemical activities, which are catalase, oxidase, MR, VP, indole, hydrogen sulphide, urea, starch, gelatin, cellulose, citrate, reduction of nitrate, casein, and lipolytic reactions, were analysed. The 16S rRNA gene amplification and DNA sequencing analysis were also completed for molecular identification purposes.
Isolation of PPFM bacteria
PPFM is commonly found in plant structures, especially in leaves. Even though PPFM colonised the surface leaves of a plant, their population size may be different, as can be seen in Table 1, which shows significant differences in PPFM population between 3 types of ulam leaves. Based on the table, the total number mean log CFU/g of PPFM colonies at the leaf surface of C. caudatus is 4.44 ± 0.07, followed by O. javanica, 3.75 ± 0.20, and M. lunu-ankenda, 3.22 ± 0.07. C. caudatus leaves have the highest number of PPFM, followed by O. javanica leaves, and the lowest is on M. lunu-ankenda leaves. The mean number of PPFM from O. javanica (3.75 ± 0.20 log CFU/g) leaves was similar to the PPFM at red clover leaves (3.82 log CFU/g), while the mean number of PPFM at M. lunu-ankenda leaves (3.22 ± 0.07 log CFU/g) was similar to the same leaves (3.18 log CFU/g), as reported by Omer et al.53. However, the mean number of PPFM at C. caudatus leaves (4.44 ± 0.07 log CFU/g) agrees with Dourado et al.,54 who claim that the population of PPFM bacteria is in the range of 4.00-6.00 log CFU/g of plant tissue.
Table (1):
The number of PPFM bacteria colonies (log CFU/g) at the leaf surface of ulam
No. |
Sample |
Total colony (CFU/g) |
Log CFU/g |
Mean log CFU/g |
|---|---|---|---|---|
1 |
C. caudatus |
3.2 × 104 |
4.5 |
4.4 ± 0.1a |
2.7 × 104 |
4.4 |
|||
2.4 × 104 |
4.4 |
|||
2 |
O. javanica |
9.4 × 103 |
4.0 |
3.8 ± 0.2b |
5.0 × 103 |
3.7 |
|||
3.9 × 103 |
3.6 |
|||
3 |
M. lunu-ankenda |
1.8 × 103 |
3.2 |
3.2 ± 0.1c |
1.9 × 103 |
3.3 |
|||
1.4 × 103 |
3.1 |
Note: Mean values ± SD with different letters in the same column have significance different (p < 0.05)
The diverse CFU/g number of PPFM on plant leaves may differ due to several factors, for example, different species of plants as well as seasonal change.30,53 Further details reported by Hunter et al.55 are the plant species, gross plant morphology, the position and height of leaves, leaf age, the distribution of stomata, trichomes, and other leaf surface appendages on the waxes surface, together with distinct epidermis and layer,56 which also play roles for PPFM to colonise the leaf surfaces. For instance, the number of culturable bacteria obtained from broad-leaf plants is higher as compared to grasses or waxy broad-leaf plants.57
In other words, the morphology of the leaf surface may influence the ability of PPFM to make the leaf surface their resident place in the fluctuating environment.57 This is because the thick waxy cuticles limit the diffusion of nutrients and inhibit the wetting of the leaf surface.57 If water molecules fall on the thick waxy leaves, they will stay in tightly formed droplets rather than spreading over the surface,58 giving the PPFM a low chance of obtaining the water source. Hence, those explanations likely confirm the reason for different population numbers of PPFM at different types of ulam leaves. This information may aid in obtaining various species of PPFM or providing ulam selection for consumption preference.
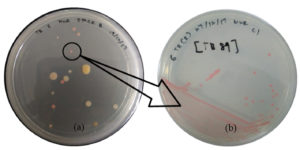
Figure 1. (a) The microbes including PPFM colonies (in black circle) on the surface of modified Pseudomonas agar (MPA) and (b) single PPFM colony culture
During the isolation step, Figure 1 (a) shows the various microbes that are present at the surfaces of ulam leaves, including PPFM bacteria colonies (in a black circle) that grow on the MPA, while Figure 1 (b) shows the pure culture streak of a single PPFM colony that has been transferred from the mixture culture plate. PPFM is expected to be isolated from the ulam leaves based on the fact that they exist in the plant’s leaves abundantly53 since leaves can support a large and complex population of bacteria.30
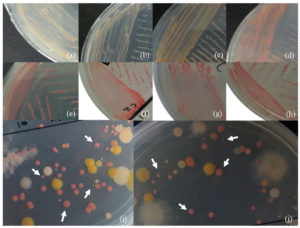
The distinctive pink colour of PPFM colonies can be present in different ranges or intensities. Based on previous studies, they described their PPFM colonies’ colour as light or pale pink, dark or deep pink, pink, and orange-pink.59,60 Therefore, a few examples of different pink variations of PPFM colonies collected from the ulam have been presented in Figure 2, including their size differences, similar to findings reported by Kumar & Lee,35 Vadivukkarasi,59 and Kassem et al.60
Figure 2. Examples of the pink colour of the PPFM colony – (a,b) light or pale pink; (c, d) orange-pink; (e, f) pink; (g, h ) dark or deep pink; (i, j ) and their variety of sizes (white arrow)
PPFM is a member of the genus Methylobacterium that belongs to the proteobacterial subgroup (class) Alpha-proteobacteria, order Rhizobiales, and family Methylobacteriaceae. The important characteristic of identifying PPFM is their ability to oxidise methanol due to the presence of the methanol dehydrogenase (mxaF) gene in their gene’s code.24,61 Physically, their pink colony feature is another unique characteristic of PPFM because of carotenoid’s existence in their cell structure. This carotenoid element is important to allow them to survive in extreme conditions such as excessive light exposure and radiation.35
Morphology and biochemical activities
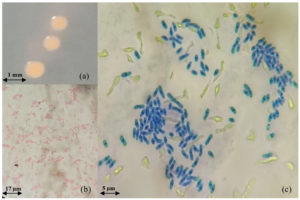
Generally, the PPFM colonies’ diameter is approximately 1.03 ± 0.06 mm with a reddish-pink colour, circular form, entire margin, raised elevation, butyrous consistency surface, and opaque colonies, as shown in Figure 3 (a). Those morphologies are identical to the morphology of PPFM described by Hirano and Upper62 and Uy et al.63 In detail, the Gram staining type (positive or negative), size, shape, mobility, and 14 biochemical tests of the selected isolates (OJ4, OJ154, and ML8) were recorded. Based on those tables, all 3 isolates belong to the Gram-negative group and are motile with rod shapes, as can be seen similarly in Figures 3 (b) and (c). Moreover, in Figure 3 (c), the isolated cell structure was stained by methylene blue solution on a glass slide before being covered with a glass cover slip. In this study, instead of relying on Gram staining, simple methylene blue staining was added for morphology examination because this assay shows the brilliant colour of bacteria cells, making them stand out against their background as this alkaline cationic dye attracted to slightly negatively charged bacteria cells for a more presentable image.64-66 Therefore, only the live cell was stained, and the unstained thing shown in the image was due to the glass reflection during the microscopy observation and photography process. Moreover, only one photograph was shown as a representative among the 3 selected strains, OJ4, OJ154, and ML8, since they have the same or similar physical structure. The size (l × w) in µm units of OJ4 is 4.3 ± 1.1 × 1.6 ± 0.6, while OJ154 is 5.4 ± 0.2 × 1.2 ± 0.1, and ML8 isolates are 3.5 ± 0.7 × 1.0 ± 0.0.
Figure 3. (a) PPFM colonies on the surface of selective agar after 6 days incubation (b) Gram staining (mag.: 400×) (c) Methylene blue cell staining (mag.: 1000×)
All isolates show positive urease, catalase, and oxidase activities, while none of the isolates can degrade starch, gelatine, or cellulose, as well as glucose fermentation (MR test) and metabolism actions (VP test), producing indole and hydrogen sulphide gaseous. In addition, only OJ154 demonstrates positive casein hydrolysis and nitrate reduction activities. However, only ML8 can utilise citrate, but not in lipid degradation. Particularly, Gram stains were done on all isolates to confirm their groups since bacteria have 2 large groups – Gram-positive and Gram-negative. These 2 groups refer to the peptidoglycan layer in the cell wall of bacteria, where the Gram-positive group has a thick peptidoglycan layer and the Gram-negative group has a thin peptidoglycan layer with an additional outer membrane composed of lipopolysaccharide. The gram-positive group shows a purple colour as a result of crystal violet stains that remain in their thick peptidoglycan layer, while the Gram-negative group shows a red colour from counterstaining safranin in their thin peptidoglycan layer during the Gram staining procedure.67 Thus, all the collected isolates belong to Gram-negative, as all of them show a red colour after the Gram staining process.
From a biochemical perspective, all 3 isolates produce the urease enzyme to breakdown urea substances into ammonia, carbon dioxide, and water, as well as the catalase enzyme to breakdown hydrogen peroxide into water and oxygen (bubble formation) during the catalase test and the oxidase enzyme for cell respiration, according to Faddin.45 Besides, most of the isolates have a lipase enzyme to change the tributyrin (lipid) structure into glycerol and butyric acid (a fatty acid), showing transparent, clear zone formation around colonies on tributyrin-based (glycerol tributyrate) agar.68,69
On the other hand, none of them expresses the α-amylase enzyme to hydrolyse starch, including the gelatinase and cellulase enzymes, since there is no gelatine liquefaction activity or cellulose compound degradation.45,70 Also, they can’t ferment glucose because there is no production of hydrogen ions and acetoin, no indole production from the activity of tryptophan breakdown, indicating no tryptophanase enzyme present, and no production of hydrogen sulphide gaseous, showing no reduction of sulphur-containing amino acids or through the reduction of inorganic sulphur compounds protein degradation by the enzyme cysteine desulfurase/desulfhydrase.45,71,72
In addition, only OJ154 is capable of hydrolysing casein, demonstrating proteinase (caseinase) and nitrate reductase enzymes present in its cell for nitrate reduction.45,73,74 Only ML8 has the citritase or citrase demolase enzyme to utilise citrate.45 From all the biochemical activity data, various enzymes produced by isolates are essential to digest macromolecules into smaller molecules that can be used as nutrients for their development and to maintain their survival.75
In summary, the compilation of microscopic observation and biochemical activity characteristics of all isolates from ulam leaves shows similar PPFM characteristics as defined by Ito and Izuka,76 Green,36 Wang et al.,77 Green and Ardley,78 and Bijlani et al.79 in Table 2. Hence, those PPFM isolates in this study most probably belong to the Methylobacterium genus.
Table (2):
The characteristic description of Methylobacterium and Methylorubrum genera and a few species of them from previous studies
No. |
Characteristic |
Methylobacterium sp.36 |
M. radiotolerans76 |
OJ4 |
ML8 |
M. ajmalii79 |
Methylorubrum sp.78 |
M. salsuginis77 |
OJ154 |
|---|---|---|---|---|---|---|---|---|---|
1. |
GS |
Negative |
Negative |
Negative |
Negative |
Negative |
Negative/variable asporogenous |
Negative |
Negative |
2. |
Size (µm)
(l×w) |
1.0 – 8.0 × 0.8 – 1.0 µm |
1.4 – 2.5 × 0.6 – 0.8 µm |
4.3 ± 1.1 × 1.6 ± 0.6 µm |
3.5 ± 0.7 × 1.0 ± 0.0 µm |
2.2 – 3.2 × 1.6 – 1.8 µm |
1.0 – 10.0 × 0.5 – 1.5 µm |
4.0 – 8.0 × 1.0 – 1 .5 µm |
5.4 ± 0.2 × 1.2 ± 0.1 µm |
3. |
Shape |
Rod |
Rod |
Rod |
Rod |
Rod |
Rod |
Rod |
Rod |
4. |
Motility |
Motile |
Motile |
Motile |
Motile |
Motile |
Motile (most strain) |
Motile |
Motile |
5. |
SH |
– (most strain) |
+ (slight) |
– |
– |
ND |
– (most strain) |
+ |
– |
6. |
CH |
– (most strain) |
– |
– |
– |
ND |
– (most strain) |
ND |
+ |
7. |
GH |
– (most strain) |
– |
– |
– |
ND |
– (most strain) |
– |
– |
8. |
CD |
– (most strain) |
ND |
– |
– |
ND |
– (most strain) |
ND |
– |
9. |
CUT |
+ (some strain) |
+ |
– |
+ |
ND |
– (most strain) |
+ |
– |
10. |
LA |
+ (some strain) |
ND |
+ |
– |
ND |
+ (some strain) |
ND |
+ |
11. |
UT |
+ |
+ |
+ |
+ |
ND |
+ |
+ |
+ |
12. |
MRT |
– |
ND |
– |
– |
ND |
– |
– |
– |
13. |
VPT |
– |
ND |
– |
– |
ND |
– |
– |
– |
14. |
NRT |
+ (some strain) |
+ |
– |
– |
ND |
+ (some strain) |
+ |
+ |
15. |
CA |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
16. |
IP |
– (except M. thiocyanatum) |
– |
– |
– |
ND |
+ (some strain) |
– |
– |
17. |
HSP |
– |
+ |
– |
– |
ND |
– (most strain) |
– |
– |
18. |
OT |
+ (often weak) |
+ |
+ |
+ |
+ |
+ (often weak) |
ND |
+ |
Note: (+) positive reaction; (-) negative reaction; ND = no data available, GS: Gram staining, l: Length, w: Width, MO: Microscopic observation, SH: Starch hydrolysis, CH: Casein hydrolysis, GH: Gelatin hydrolysis, CD: Cellulose degradation, CUT: Citrate utilization test, LA: Lipolytic activity, UT: Urease test, MRT: Methyl red test, VPT: Voges-Proskauer test, NRT: Nitrate reduction test, CA: Catalase activity, IP: Indole production, HSP: Hydrogen sulphide production, OT: Oxidase test
Further tests, which are molecular identifications, were done to identify the species of the isolates. The 16S rRNA sequence analysis of the selected isolates acted as a complement to verify their closest related species.
Molecular identification
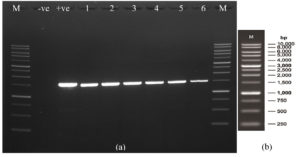
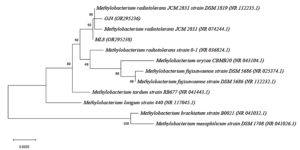
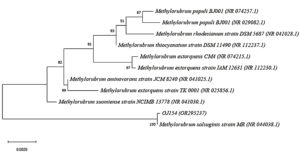
For species confirmation, the 3 selected isolates undergo a deep gene profile. Figures 4 (a) and (b) demonstrate the band of the 16S rRNA gene after being amplified from the 3 selected isolates’ genes at 1500 base pairs. Then, the 16S rRNA partial gene sequences of the 3 isolates were matched with BLAST from the NCBI database for species identification. According to the 16S rRNA sequence (Table 3), the most closely related (99% similarity) bacteria species to the isolate OJ4 is M. radiotolerans JCM 2831 strain DSM 1819, while OJ154 is M. salsuginis strain MR. Similar to isolate OJ4 with different strains, the most closely related (99% similarity) bacteria species of isolate ML8 is M. radiotolerans JCM 2831. Additionally, the phylogenetic tree based on the 16S rRNA sequence of members of the genus Methylobacterium shows the location of each isolate (Figures 5 and 6). Those phylogenetic trees display the close relationship of OJ4 with M. radiotolerans JCM 2831 strain DSM 1819, as well as ML8 with M. radiotolerans JCM 2831. Differently, OJ154 has a close relationship with M. salsuginis strain MR.
Table (3):
The 16S rRNA sequence of isolates based on the BLAST database
| Isolate name | Length of 16S rRNA gene sequenced (bp) | Most closely related organism | |||
|---|---|---|---|---|---|
| Species | Accession description | % Gene identity | % Query coverage | ||
| OJ4 | 1418 | M. radiotolerans JCM 2831 strain DSM 1819 | NR_112235.1 | 99% | 100% |
| OJ154 | 1411 | M. salsuginis strain MR | NR_044038.1 | 99% | 100% |
| ML8 | 1411 | M. radiotolerans JCM 2831 | NR_074244.1 | 99% | 100% |
Figure 4. (a) The gel photo of the band of the 16S rRNA gene of the isolate after amplification at 1500 base pairs – band: 5 (OJ4), 6 (OJ154), 3 (ML8) (b) base pair indicator
Figure 5. Neighbor-joining phylogenetic tree based on 16S rRNA gene sequence shows the relationship of the samples [OJ4 (OR295236)] and [ML8 (OR295238)] with members of the genus Methylobacterium. Bootstrap values from 1000 replications are shown at branch points. Bar, 0.002 substitution per site
Figure 6. Neighbor-joining phylogenetic tree based on 16S rRNA gene sequence shows the relationship of the sample [OJ154 (OR295237)] with members of the genus Methylobacterium. Bootstrap values from 1000 replications are shown at branch points. Bar, 0.002 substitution per site
As discussed by Green and Ardley,78 some genera of Methylobacterium have been reclassified into a new genus, namely Methylorubrum, due to a few dissimilarities. For example, most species of Methylorubrum gen. nov. can utilise methylamine as the sole source of carbon and energy, whereas most species belonging to Methylobacterium cannot. Therefore, OJ154 falls into this newly classified genus, Methylorubrum. As mentioned previously, all the isolates are considered members of the Methylobacterium genus via microscopic examination and a few biochemical tests. However, a recent study discovered a new genus (Methylorubrum), which is similar to the Methylobacterium characteristic, raising the possibility that some of the isolates also belong to the Methylorubrum genus. Based on Table 3, OJ4 and ML8 show similar characteristics of M. radiotolerans, whereas OJ154 resembles M. salsuginis. Though they are closely related to the same species, each of them has a few different biochemical activities. This phenomenon may occur due to the different strains of the same species.80
Additionally, further chemotaxonomic characterisation of the species was not performed because the main objective of this study was to focus on the food product application (edible bacteria pigment). The species of Methylobacterium have been known ubiquitously everywhere, including at international space stations.37,79 In most terrestrial plants, of which there are more than 70 species, they can be found actively colonising the roots, branches, seeds, and leaves, as reported by Irvine et al.,14 Mizuno et al.,30 Iguchi et al.,81 and Dourado et al.54 and live dominantly on the leaf surface.57 They obtain their carbon source from various carbon sources released by plants that leach from the cuticle and the volatile carbon substrates, mainly byproduct methanol released from cell wall metabolism via stomata.11 Therefore, the taxonomic characterisation procedures that have been performed are to prove their presence on the surface of selected ulam leaves even after washing them with sterile distilled water during the isolation process.
The PPFM bacteria have been isolated from ulam, and the highest population of them can be found in C. caudatus leaves, followed by O. javanica and M. lunu-ankenda. Selected isolate, namely ML8, can be considered the species of M. radiotolerans JCM 2831, similar to isolate OJ4 species but a different strain, M. radiotolerans JCM 2831 strain DSM 1819, while isolate OJ154 was characterised by the species of M. salsuginis strain MR based on their morphology, biochemical activities, and 16S rRNA gene sequence characteristics.
ACKNOWLEDGMENTS
The authors express their gratitude to Universiti Putra Malaysia for the provision of essential facilities and funding.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was funded by the Putra Grant-Putra Graduate Initiative (GP-IPS/2017/9588200) and Graduate Research Fellowship.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Miglio C, Chiavaro E, Visconti A, Fogliano V, Pellegrini N. Effects of different cooking methods on nutritional and physicochemical characteristics of selected vegetables. J Agric Food Chem. 2008;56(1):139-147.
Crossref - Cheng SH, Barakatun-Nisak MY, Anthony J, Ismail A. Potential medicinal benefits of Cosmos caudatus (Ulam Raja): A scoping review. J Res Med Sci. 2015;20(10):1000-1006.
Crossref - Bachok MF, Yusof BNM, Ismail A, Hamid AA. Effectiveness of traditional Malaysian vegetables (ulam) in modulating blood glucose levels. Asia Pacific J Clin Nutr. 2014;23(3):369-376.
Crossref - Chan EWC, Wong SK, Chan HT. Ulam Herbs of Oenanthe javanica and Cosmos caudatus: An overview on their Medicinal Properties. Journal of Natural Remedies. 2017;16(4):137-147.
Crossref - Fatimah AMZ, Norazian MH, Rashidi O. Identification of carotenoid composition in selected “ulam” or traditional vegetables in Malaysia. Int Food Res J. 2012;19(2):527-530.
- You YX, Shahar S, Haron H, Yahya H, Din NC. Relationship between traditional Malaysian vegetables (ulam) intake and cognitive status among middle-aged adults from low cost residential areas. Jurnal Sains Kesihatan Malaysia. 2019;17(S1):139-148.
Crossref - Randhawa MA, Khan AA, Javed MS, Sajid MW. Green leafy vegetables: A health promoting source. In: Watson RR, ed. Handbook of Fertility. Academic Press. 2015:205-220.
Crossref - Lu CL, Li XF. A review of Oenanthe javanica (Blume) DC. as traditional medicinal plant and its therapeutic potential. Evid Based Complement Alternat Med. 2019;2019:6495819.
Crossref - Mediani A, Abas F, Khatib A, Tan CP. Cosmos caudatus as a potential source of polyphenolic compounds: optimisation of oven drying conditions and characterisation of its functional properties. Molecules. 2013;18(9):10452-10464.
Crossref - Eliaser EM, Ho JH, Hashim NM, Rukayadi Y, Ee GCL, Razis AFA. Phytochemical Constituents and Biological Activities of Melicope lunu-ankenda. Molecules. 2018;23(10):2708.
Crossref - Prajapati RR, Jhala YK, Vyas RV. In vitro study of plant growth promoting methylotrophic bacterial consortium as a plant probiotics for paddy. Int J Curr Microbiol Appl Sci. 2017;6(5):2608-2626.
Crossref - Fedorov DN, Doronina NV, Trotsenko YA. Phytosymbiosis of aerobic methylobacteria: New facts and views. Microbiology. 2011;80(4):443-454.
Crossref - Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME. The Expanding World of Methylotrophic Metabolism. Ann Rev Microbiol. 2009;63(1):477-499.
Crossref - Irvine IC, Brigham CA, Suding KN, Martiny JB. The abundance of pink-pigmented facultative methylotrophs in the root zone of plant species in invaded coastal sage scrub habitat. PloS one. 2012;7(2):e31026-e31026.
Crossref - Singh M, Kumar A, Singh R, Pandey KD. Endophytic bacteria: A new source of bioactive compounds. 3 Biotech. 2017;7:1-14.
Crossref - Narayanan Z, Glick BR. Secondary metabolites produced by plant growth-promoting bacterial endophytes. Microorganisms. 2022;10(10):2008.
Crossref - Kusari S, Pandey SP, Spiteller M. Untapped mutualistic paradigms linking host plant and endophytic fungal production of similar bioactive secondary metabolites. Phytochemistry. 2013;91:81-87.
Crossref - Sharma H, Rai AK, Dahiya D, Chettri R, Nigam PS. Exploring endophytes for in vitro synthesis of bioactive compounds similar to metabolites produced in vivo by host plants. AIMS Microbiol. 2021;7(2):175-199.
Crossref - Puri SC, Verma V, Amna T, Qazi GN, Spiteller M. An endophytic fungus from Nothapodytes foetida that produces camptothecin. J Nat Prod. 2005;68(12):1717-1719.
Crossref - Kusari S, Zuhlke S, Spiteller M. An endophytic fungus from Camptotheca acuminata that produces camptothecin and analogues. J Nat Prod. 2009;72(1):2-7.
Crossref - Kusari S, Zuhlke S, Spiteller M. Effect of artificial reconstitution of the interaction between the plant Camptotheca acuminata and the fungal endophyte Fusarium solani on camptothecin biosynthesis. J Nat Prod. 2011;74(4):764-775.
Crossref - Shweta S, Zuehlke S, Ramesha BT, et al. Endophytic fungal strains of Fusarium solani, from Apodytes dimidiata E. Mey. ex Arn (Icacinaceae) produce camptothecin, 10-hydroxycamptothecin and 9-methoxycamptothecin. Phytochemistry. 2010;71(1):117-122.
Crossref - Sarjono PR, Putri LD, Budiarti CE, et al. Antioxidant and antibacterial activities of secondary metabolite endophytic bacteria from papaya leaf (Carica papaya L.). In: IOP Conf Ser: Mater Sci Eng. 2019:509(1):012112.
Crossref - Balachandar D, Raja P, Sundaram SP. Genetic and metabolic diversity of pink-pigmented facultative methylotrophs in phyllosphere of tropical plants. Braz J Microbiol. 2008;39(1):68-73.
- Jones SW, Karpol A, Friedman S, Maru BT, Tracy BP. Recent advances in single cell protein use as a feed ingredient in aquaculture. Curr Opin Biotechnol. 2020;61:189-197.
Crossref - Feinberg LF, Marx CJ, Wall MA, Smith DR, Pujol-Baxley CJ, Mcavoy BD. Heterologous expression of taurine in microorganisms. U.S. Patent No. 11,326,171. 2022.
- Hardy RW, Patro B, Pujol-Baxley C, Marx CJ, Feinberg L. Partial replacement of soybean meal with Methylobacterium extorquens single-cell protein in feeds for rainbow trout (Oncorhynchus mykiss Walbaum). Aqua Res. 2018;49(6):2218-2224.
Crossref - EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA), Turck D, Bresson J-L, Burlingame B, et al. Safety of pyrroloquinoline quinone disodium salt as a novel food pursuant to Regulation (EC) No 258/97. EFSA Journal. 2017;15(11)
Crossref - Madhaiyan M. Molecular Aspects, Diversity and Plant Interaction of Facultative Methylotrophs Occurring in Tropical Plants. [Doctoral dissertation]. Coimbatore, Lawley Road: Tamil Nadu Agricultural University. 2003.
- Mizuno M, Yurimoto H, Yoshida N, Iguchi H, Sakai Y. Distribution of pink-pigmented facultative methylotrophs on leaves of vegetables. Biosci Biotechnol Biochem. 2012;76(3):578-580.
Crossref - Sanjee SA, Karim M. Microbiological quality assessment of frozen fish and fish processing materials from Bangladesh. Int J Food Sci. 2016;2016:8605689.
Crossref - Kaparullina EN, Doronina NV, Mustakhimov II, Agafonova NV, Trotsenko YA. Biodiversity of aerobic methylobacteria associated with the phyllosphere of the southern Moscow region. Microbiology. 2017;86(1):113-118.
Crossref - Tortora GJ, Funke BR, Case CL, Warner BB, Derek W. Microbiology: An Introduction. Pearson. 2019.
- McManus JR. Characterization of Pink-Pigmented Facultative Methylotrophic Bacteria Isolated from Oil Reservoirs. [Master dissertation]. Stillwater, Oklahoma: Oklahoma State University. 1987.
- Kumar R, Lee AC. Isolation and characterization of pink-pigmented, facultative methylotrophic (PPFM) bacteria from leaves of neem, Azadirachta indica A. Juss. Philipp J Syst Biol. 2009;3(1).
- Green PN. Methylobacterium. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, ed. The Prokaryotes: A Handbook on The Biology of Bacteria: Proteobacteria: Alpha and Beta Subclasses. Springer. 2006:257-265.
Crossref - Barrow GI, Feltham RKA. Cowan and Steel’s Manual for the Identification of Medical Bacteria. Cambridge University Press. 1993.
Crossref - Engelkirk PG, Duben-Engelkirk J. Burton’s Microbiology for the Health Sciences. Wolters Kluwer Health/Lippincott Williams & Wilkins. 2011.
- United States. Food and Drug Administration. Division of Microbiology. Bacteriological Analytical Manual of the Division of Microbiology, Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration. Association of Official Analytical Chemists. 1984.
- Laxmi PVJ, Reddy NN, Rajsekaran C, Kumari D, Mitra B. Standardization of cultivation parameters for the extraction of carotenoid from pink pigmented facultative methylotrophic (PPFM) bacteria. Asian J Pharm Clin Res. 2012;5(2):52-57.
- Pattanashetti SR. Isolation and Characterization of Pink Pigmented Facultative Methylotrophs from Coleus Forskohlii and Their Influence on Growth and Tuber Yield. [Doctoral dissertation]. GKVK, Bangalore: University of Agricultural Sciences. 2012.
- Atlas RM. Handbook of Microbiological Media. CRC Press. 2010.
Crossref - Aneja KR. Experiments in Microbiology Plant Pathology and Biotechnology. New Age International Publishers. 2010.
- Tille, PM. Bailey and Scott’s Diagnostic Microbiology. Elsevier Mosby. 2014.
- Faddin JFM. Biochemical Tests for Identification of Medical Bacteria. Williams & Wilkins Company. 1980.
- Public Health England. UK Standards for Microbiology Investigations Oxidase Test. England: Standards Unit, National Infection Service, Public Health; 2019. https://www.gov.uk/government/publications/smi-tp-26-oxidase-test. Accessed October 27, 2023.
- Gaby WL, Hadley C. Practical laboratory test for the identification of Pseudomonas aeruginosa. J Bacteriol. 1957;74(3):356-358.
Crossref - Reysenbach AL, Longnecker K, Kirshtein J. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl Environ Microbiol. 2000;66(9):3798-3806.
Crossref - Anggraini L, Marlida Y, Wizna W, et al. Molecular identification and phylogenetic analysis of GABA-producing lactic acid bacteria isolated from indigenous dadih of West Sumatera, Indonesia. F1000Research. 2019;7:1663.
Crossref - Vingron, M, Stoye J, Luz H, Bocker S. Algorithms for Phylogenetic Reconstructions. [Lecture, Notes and Exercises]. Berlin, Rod: 2003. Accessed October 28, 2023.
- Munjal G, Hanmandlu M, Srivastava S. Phylogenetics algorithms and applications. In: Hu Y-C, Tiwari S, Mishra KK, Trivedi MC, ed. Ambient Communications and Computer Systems: RACCCS-2018. Springer. 2019:187-194.
Crossref - Hong Y, Guo M, Wang J. ENJ algorithm can construct triple phylogenetic trees. Molecular Therapy-Nucleic Acids. 2021;23:286-293.
Crossref - Omer ZS, Tombolini R, Gerhardson B. Plant colonization by pink-pigmented facultative methylotrophic bacteria (PPFM). FEMS Microbiol Ecol. 2004;47(3):319-326.
Crossref - Dourado MN, Aline ACN, Santos DS, Araujo WL. Biotechnological and Agronomic Potential of Endophytic Pink-Pigmented Methylotrophic Methylobacterium spp. BioMed Res Int. 2015:909016.
Crossref - Hunter PJ, Hand P, Pink D, Whipps JM, Bending GD. Both leaf properties and microbe-microbe interactions influence within-species variation in bacterial population diversity and structure in the lettuce (Lactuca species) phyllosphere. Appl Environ Microbiol. 2010;76(24):8117-8125.
Crossref - Hirano SS, Upper CD. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-A Pathogen, Ice Nucleus, and Epiphyte. Microbiol Mol Biol Rev. 2000;64(3):624-653.
Crossref - Lindow SE, Brandl MT. Microbiology of the phyllosphere. Appl Environ Microbiol. 2003;69(4):1875-1883.
Crossref - Beattie G, Lindow S, Hecht-Poinar E, Elliott VJ. Leaf surface waxes and the process of leaf colonization by microorganisms. In: Lindow SE, Hecht-Poinar EI, Elliott VJ. ed. Phyllosphere Microbiology. American Phytopathological Society (APS Press); 2002:3-26.
- Vadivukkarasi P. Studies on Isolation Characterization and Applications of Pink Pigmented Facultative Methylotrophs PPFMs from Diverse Environments of Chennai India. [Doctoral dissertation]. Chennai, Tamil Nadu: University of Madras. 2013.
- Kassem MM, Hauka FI, Afify AH. Nour El-Din M, Omara AA. Isolation and identification of some PPFMs bacterial isolates and their potentiality as biofertilizers and biocontrol agents to Rhizoctonia solani. J Agric Chem Biotechnol. 2013;4(2):79-92.
Crossref - Raghavendra J, Santhosh GP. Utilization of different carbon substrates by native pink pigmented facultative methylotrophs isolated from direct seeded rice. J Pharmacogn Phytochem. 2019;8(4):2245-2247.
- Hirano SS, Upper CD. Bacterial community dynamics. In: Andrews JH, Hirano SS, ed. Microbial Ecology of Leaves. Springer Science & Business Media. 1991:271-294.
Crossref - Uy MM, Uy J, Carvajal TM, Castro CZR, Ho HT, Lee AC. Pink pigmented facultative methylotrophic (PPFM) bacteria isolated from the hair scalp and nasal cavity. Phillipp J Syst Biol. 2013;7:13-21.
- Mehrotra RS, Sumbali G. Principles of Microbiology. Tata McGraw-Hill Education. 2009.
- Kiuchi K. Rapid alkaline methylene blue supravital staining for assessment of anterior segment infections. Clin Ophthalmol. 2016;10:1971-1975.
Crossref - Procop GW, Church D, Hall G, et al. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. Jones & Bartlett Publishers. 2020.
- Alexander SK, Niles MJ, Strete D. Laboratory Exercises in Organismal and Molecular Microbiology. Mcgraw-Hill. 2004.
- Lee SH, Park DH. Isolation and physiological characterization of Bacillus clausii SKAL-16 isolated from wastewater. J Microbiol Biotechnol. 2008;18(12):1908-1914.
Crossref - Carrazco-Palafox J, Rivera-Chavira BE, Ramirez-Baca N, Manzanares-Papayanopoulos LI, Nevarez-Moorillon GV. Improved method for qualitative screening of lipolytic bacterial strains. MethodsX. 2018;5:68-74.
Crossref - Gupta P, Samant K, Sahu A. Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int J Microbiol. 2012;2012:578925.
Crossref - McDevitt, S. Methyl red and Voges-Proskauer test protocols. American Society for Microbiology. 2009.
- GroBhennig S, Ischebeck T, Gibhardt J, Busse J, Feussner I, Stulke J. Hydrogen sulfide is a novel potential virulence factor of Mycoplasma pneumoniae: Characterization of the unusual cysteine desulfurase/desulfhydrase HapE. Mol Microbiol. 2016;100(1):42-54.
Crossref - Salisbury WA, Likos JJ. Hydrolysis of casein: A differential aid for the identification of Serratia marcescens. J Clin Pathol. 1972;25(12):1083-1085.
Crossref - Hussein NNA, Ibrahim AI, Kamar FH, Nechifor AC. Caseinase production and media optimization from Bacillus subtilis. Revista de Chimie. 2020;71(11):1-9.
Crossref - Cezairliyan B, Ausubel FM. Investment in secreted enzymes during nutrient-limited growth is utility dependent. Proc Natl Acad Sci. 2017;114(37). E7796-E7802.
Crossref - Ito H, Iizuka H. Taxonomic studies on a radio-resistant Pseudomonas: Part XII. Studies on the microorganisms of cereal grain. Agric Biol Chem. 1971;35(10):1566-1571.
Crossref - Wang X, Sahr F, Xue T, Sun B. Methylobacterium salsuginis sp. nov., isolated from seawater. International J Sys Evolut Microbiol. 2007;57(8):1699-1703.
Crossref - Green PN, Ardley JK. Review of the genus Methylobacterium and closely related organisms: A proposal that some Methylobacterium species be reclassified into a new genus, Methylorubrum gen. nov. Int J Sys Evol Microbiol. 2018;68(9):2727-2748.
Crossref - Bijlani S, Singh NK, Eedara VV, et al. Methylobacterium ajmalii sp. nov., Isolated from the International Space Station. Front Microbiol. 2021;12:534.
Crossref - Baron E. Medical Microbiology, 4th Ed. The University of Texas Medical Branch at Galveston. 1996:1-21.
- Iguchi H, Yurimoto H, Sakai Y. Interactions of methylotrophs with plants and other heterotrophic bacteria. Microorganisms. 2015;3(2):137-151.
Crossref
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.