ISSN: 0973-7510
E-ISSN: 2581-690X
Microorganisms such as Pseudomonas aeruginosa have always been adaptable in surviving the harsh environment such as antimicrobial agents via the quorum sensing (QS) mechanism. Studies have shown that quorum sensing mechanism cases have been highly associated with foodborne illnesses. Since synthetic compounds such as azithromycin (AZM) are reported to have detrimental effects on human, using medicinal local plants have been gaining attention as an anti-quorum agent. The aim of this study was to determine the anti-quorum sensing activity of the Curcuma xanthorrhiza Roxb. extract against P. aeruginosa ATCC35554 quorum sensing system including swarming motility, pyocyanin production and biofilm formation. The results indicated that the extract required a high concentration to inhibit and kill the P. aeruginosa with minimum inhibition concentration (MIC) and minimum bactericidal concentration (MBC) values of 200 and >700 mg/mL, respectively. Thus, anti-quorum sensing assays were done in concentration up to 200 mg/mL. The inhibition of quorum sensing activity of C. xanthorrhiza Roxb. extract on P. aeruginosa quorum sensing was concentration dependent manner. At 200 mg/mL of the extract exhibited 72.12% reduction of swarming motility, 84.30% inhibition of the pyocyanin production and 78.35% reduction in the biofilm formation. In conclusion the crude extract of C. xanthorrhiza Roxb. extract has ability to reduce the virulence factors; swarming motility, pyocyanin production and biofilm formation regulated by quorum sensing. Thus, the extract C. xanthorrhiza Roxb. extract has anti-quorum sensing or quorum quenching activity.
Anti-quorum sensing, Curcuma xanthorrhiza Roxb., Pseudomonas aeruginosa, swarming motility, pyocyanin production, biofilm formation.
The global food safety and the economy have been reported to be seriously affected by the increasing foodborne illnesses and food spoilage1,2. Contamination by the foodborne pathogens have led to numerous cases of diarrhea, vomiting, abdominal pain and even deaths3-5. While urbanized countries such as the United States and the United Kingdom have a good statistical reports on the phenomenon6, less developed countries such as Malaysia are not as able to efficiently tackle the issue due to the lack of incidents reported7,8.
In order to combat and prevent the foodborne contaminations, antimicrobial agents used often possess a selective toxicity and targets the difference between the microorganism metabolism and the human cell’s’ structures and features9,10. However, the frequent usages of these products such as antibiotics have led to an emergence of antibiotic resistant bacteria strain11. Therefore, to reduce the antibiotic dependencies, alternative stratagems such as quorum sensing have been researched12,13.
Quorum sensing (QS) is a bacteria cell-to-cell communication mechanism used to determine the bacterial physiology including the local population density as well as adapting to the harsh and ever-changing environment such as antibacterial agents innate immune responses14-16. QS is a type of bacterial communication systems that allow determination of the bacterial physiology via the production of diffusible signaling molecules known as autoinducers (AI) such as oligopeptides and N-acyl homoserine lactones in Gram-positive and Gram-negative bacteria, respectively15. QS system is greatly associated with bacterial pathogenicity and spoilage contamination on food products17. Through the production of diffusible signaling molecules known as autoinducers (AI) in Gram-positive (oligopeptides) and Gram-negative (N-acyl homoserine lactone (AHL)) bacteria15, these microorganisms are able to regulate the QS mechanism. P. aeruginosa QS systems have been reported to be highly adaptable in responding towards the external biological stresses by producing virulence factors such as pyocyanin, swarming and biofilm formation19.
Since synthetic quorum quenching compounds such as azithromycin (AZM) are reported to have detrimental effects on human, usage of natural products from plant extracts have been gaining popularity in eliminating microbial contamination on the food products20. Javanese turmeric (Curcuma xanthorrhiza Roxb.) or locally known as “temu lawak” can be found in tropical countries such as Malaysia and Indonesia21. C. xanthorrhiza Roxb. has been traditionally used for food and medicinal purposes22,23. The Javanese turmeric has been reported to have bioactive compounds including curcuminoids, camphor, geranyl acetate, zerumbone, b-curcumene, zingiberene, ar-curcumene and xanthorrhizol24. Therefore, the aim of this study is to evaluate the antimicrobial and quorum quenching activity of the ethanolic extract against P. aeruginosa in vitro.
Bacterial Strain
P aeruginosa strain used in this research was ATCC35554 obtained from the American Type Culture Collection and cultured in Pseudomonas agar. During the study, bacteria was grown at least over 12 hours in Luria Bertani (LB) broth and streaked onto a LB agar media to obtain a single P. aeruginosa colony. The culture was preserved in a sterile universal bottle at 4°C for short term holding and at -20°C for long term holding in sterile universal bottles and agar plates25,26.
Curcuma xanthorrhiza Roxb. Rhizome Sample and Extraction
A 10 month old of C. xanthorrhiza Roxb. or Javanese turmeric rhizomes were obtained from Kebun Percobaan, Cikabayan, Damaga, Bogor, Bogor Agricultural University (IPB). The rhizomes were sorted to remove all of the soil and dirt. The rhizome was then chopped manually approximately 5-7 mm. The sliced rhizomes were dried using oven drying (55°C) and powdered into 60 meshes with a grinder. C. xanthorrhiza Roxb. rhizome extraction was done as per method by Ab Halim et al.27. Briefly, 100 g of the powdered rhizome was soaked in 400 mL of ethanol for 48 hours at room temperature. The mixture was then filtered using Whatman filter No.2 and evaporated with a vacuum evaporator at 50°C to obtain a concentrated ethanolic crude extract. The ethanolic extract of C. xanthorrhiza Roxb. was preserved in an universal bottle at 4°C for further use28.
Determination of Minimum Inhibitory Concentration (MIC) and minimum Bactericidal Concentration (MBC)
Determination of MIC and MBC of C. xanthorrhiza Roxb. extract on P. aeruginosa culture were done using the standard method of Clinical and Laboratory Standards Institute (CLSI)30. Through MIC and MBC assay, the concentrations of the extract to inhibit and kill the P. aeruginosa culture, respectively, were conducted. Briefly, in the study, 10 mg/mL chlorhexidine and dimethyl sulfoxide (DMSO) were used as positive and negative controls, respectively. Since chlorhexidine have been known to for its effectiveness against vast spectrum of bacteria while DMSO is commonly used as the negative control due to the absence of antimicrobial activity. As per the CLSI30, the first two wells of the 96 well plate, were allocated for the positive and negative control for MIC assay. Meanwhile, the rest of the wells were aliquoted with the extract diluted with MH broth to obtain concentration of 10-800 mg/mL. In MBC test, 10 µL aliquot from each well was inoculated onto a sterile MH agar and incubated for 24 hours at 37°C. After the incubation, wells that shows no bacterial growth represents the minimum concentration needed to kill the P. aeruginosa.
Growth Assay
To further study the antimicrobial activity of the ethanolic extract against P. aeruginosa growth, Log10 Colony Forming Units (CFU) was done on Nutrient agar plates. The method was chosen as the bacterial count capacity can be adjusted via serial dilutions and the method allow only viable bacterial colony to be counted31. A culture (OD600 ~0.1) incubated at 37°C with shaking at 200 RPM for four hours was inoculated into sterile universal bottles containing 9 mL of LB broth. 1 mL of the extract (0-200 mg/mL) was added to the respective bottles and incubated at 37°C for 24 hours. A 100 µL of culture was aseptically spread onto Nutrient agars using a glass hockey stick before 24 hours incubation. The colonies formed were counted and the CFU/mL was calculated.
Swarming Activity Assay
Based from the antimicrobial assays, the extract concentration needed to observe the C. xanthorrhiza Roxb. quorum quenching activity against P. aeruginosa was 0-200 mg/mL. In swarming inhibition assay, approximately 0.1 OD600 P. aeruginosa culture was inoculated into universal bottles containing the extract (0-200 mg/mL) and incubated overnight at 37°C with 180 rpm shaking. The culture was then inoculated onto a swarming agar media consisting of 0.5% (w/v) agar and 8 g/L nutrient broth using a sterile toothpick32. The plates were then incubated at 37°C for 24 hours and the mean length of the swarming distance between the central of the inoculation site was determined.
Pyocyanin Production Assay
Inhibition on pyocyanin pigment production was done according to the method by King et al.33. From the swarming assay, the swarming colony was scooped from the media, cut into small pieces and added with 5 mL of saline (0.85% NaCl) in centrifuge tubes. The tubes were centrifuged at 10,000 RPM for 10 minute twice34. A 5 mL of the supernatant was mixed vigorously with 3 mL chloroform before discarding the aqueous phase34-37. The pigment was then re-extracted with 1 mL of 0.2 N HCl and the mixture was mixed vigorously to elicit a pink red solution35,38. The red pink solution was centrifuged at 8,000 RPM for 10 minutes and the relative pyocyanin concentration was measured using a spectrophotometer at OD520 with 0.2 N as blank36,38,39. The pigment concentration was then calculated by multiplying the optical density value by 17.07249,41.
Biofilm Formation Assay
The effect of the ethanolic extract of C. xanthorrhiza Roxb. against P. aeruginosa biofilm formation was done according to Varposhti et al.42. Each well of the 96-microtiter flat-well plate consist of 50 µL of the LB broth and 50 µL of the extract (0-200 mg/mL). A100 µL of the bacterial culture at 108 CFU/mL was added into each well, mixed thoroughly and incubated for 24 hours at 37°C. Post incubation, the wells were washed with pre-warmed physiological saline and let dry for 10 minutes. Then, 100 µL of the tetrazolium salt (XTT)/menadione solution was added into each well and incubated in the dark at 37°C for 4 hours. After the incubation period, the content of each well was transferred into a new 96 wells microtiter plate and the absorbance at OD490 was measured. The anti-biofilm activities of the plant extracts was calculated using the percentage mean of the optical density at 490 nm wavelength against the untreated biofilm. The decrease in biofilm formation of treated samples was compared to the untreated biofilm were determined.
Statistical Analysis
The tests were done by 2 x 2 and the results were analyzed using the MINITAB17 software. In the data analysis, the one-way variance analysis (ANOVA) and the Tukey’s test were used to determine the significant difference (P<0.05) between different concentration of plant extract used (0-200 mg/mL).
Yield of Curcuma xanthorrhiza Roxb. extract
The average crude extract yield of C. xanthorrhiza Roxb. rhizome was 4.01 ± 0.89% (Table 1). This result is comparable with the extraction yield of C. xanthorrhiza Roxb. rhizome (5.9%) obtain from Ab Halim et al.27. This difference in yield could be from variety of reasons such as the method of extraction. The method chosen for this test was chosen for its simplicity, cheap and convenience43. Soaking the powdered material in the solvent allow for the plant’s cell walls to soften and broken down, releasing the soluble phytochemicals into the solvent43.
Table (1):
Extraction yield of the Cucurma xanthorrhiza Roxb. rhizome
| Dried powder (g) | Extraction | Yield (g) | Yield 9%) |
|---|---|---|---|
| 100 100 100 |
1st 2nd 3rd |
4.98 3.22 3.82 |
4.98 3.22 3.82 |
| Average ± SD | 4.01 ± 0.89 | 4.01± 0.98 | |
SD, standard deviation
Ethanol was chosen as the solvent due to its safer and less toxic nature than acetone, methanol and other organic solvents44-46. Human liver naturally produce an enzyme known as alcohol dehydrogenase which functions to convert alcohol into acetylaldehyde as its source of energy47. However, when methanol reacts with alcohol dehydrogenase, it leads to the production of formaldehyde that is very reactive and may interact with a host molecule in the body to shut down the enzymatic pathways47. Since ethanol is a versatile solvent with a universal characteristic, it can attract both non-polar and polar compounds such as alkaloid, curcuminoid and terpenoid48,49. Ab Halim et al.27 have reported that ethanol solvent have been reported to elicit high phytochemical compounds such as phenols and tannins compared to other solvents.
Determination of minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC)
The MIC and MBC result of the C. xanthorrhiza Roxb. ethanolic extract against P. aeruginosa was 200 and >700 mg/mL (Table 2). The results indicated that a high concentration of the extract was needed to inhibit and kill the P. aeruginosa culture (200 and 800 mg/mL, respectively). In the study done by Diastuti et al.50, the MIC and MBC values of the C. xanthorrhiza Roxb. extracts on the P. aeruginosa were much lower than in this study. The difference in the concentration needed for MIC and MBC was due to the difference in the extraction methods and solvents used when isolating the phytochemical compounds from C. xanthorrhiza Roxb. rhizome. Different extraction method and solvents could have certain affinity on several phytochemicals over the other. For example, in the study by Ab Halim et al.27, ethanol extract of the C. xanthorrhiza Roxb. possesses more phytochemical compounds such as terpenes, phenols, flavonoids among others compared to the aqueous solvent. Difference of affinity in phytochemical extractions could be due to the polarity attraction from the ethanol solvent due to its universal characteristics and versatile solvent51.
Table (2):
Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of Curcuma xanthorrhiza Roxb. extract against Pseudomonas aeruginosa
C. xanthorrhiza Roxb. extract (mg/mL) |
|
|---|---|
MIC MBC |
200 >700 |
Furthermore, the high extract concentration needed for MIC and MBC could be due difference between cell wall characteristic of Gram-negative of P. aeruginosa. Mangunwardoyo et al.52 has reported that an extraction of C. xanthorrhiza Roxb. extract with aquadest, 70% ethanol and dichloromethane have also failed to exhibit zones of inhibition on the Gram-negative bacteria such as P. aeruginosa, E. coli, P. gingivalis as well as fungi C. albicans. Selim et al.53 and Mangunwardoyo et al.52 have explained that gram-negative bacteria possess a higher resistance towards antimicrobial agents as compared to the Gram-positive bacteria. Such resistance could be due to higher concentration of lipid in the cell wall54. Bacterial cell walls are commonly composed of lipopolysaccharides, lipoproteins and periplasms that are bonded to the peptidoglycans55-57. These lipopoly-saccharides in the cell wall serves as the bacteria defense system that only selectively allows foreign objects to pass through the cell wall52.
Since Gram-negative bacterial cell walls are reported to possess a non-polar characteristic, this makes polar and semi-polar derived extraction methods have a higher difficulty to permeate the cell wall58. Therefore, due to its semi-permeable outer membrane, Gram-negative bacteria are able to reduce its susceptibility against antimicrobial agents by minimizing intake of dangerous foreign substance such as antimicrobial agents59. Furthermore, P. aeruginosa outer membrane permeability is lower than other Gram-negative bacteria by 12-100 fold60. This serves as a crucial barrier for the bacteria against penetration by the antimicrobial agents as these agents will have to take a longer time to pass through P. aeruginosa cell wall58. During this time, the bacteria has ample time to intrinsically gain resistance against the agents by synergizing via its internal mechanisms such as the efflux pumps and periplasmic b-lactamases to actively pump out and/or degrade the compounds58.
Growth assay
Growth assay at log10 was done to further confirm the effect of MIC extract concentration (0-200 mg/mL) on the bacterial growth. The result showed that the C. xanthorrhiza Roxb. extract does not or have little insignificant effect on the growth of P. aeruginosa (Fig. 1). The figure shows that apart from the positive control, P. aeruginosa growth was barely affected by the C. xanthorrhiza Roxb. extract whereby the negative control (DMSO) and extract at concentration of 0-200 mg/mL showed an insignificant difference of bacterial growth ranging from 8.99 ± 0.18 to 9.73 ± 0.05 log10 CFU/mL. Similarly, Ugurlu et al.35 have also reported that concentrations up to 4 mmol/L had no effect on the growth of P. aeruginosa has shown an inhibiting effect on the bacterium quorum sensing mechanisms. Since compounds or concentration that do not kill or inhibit the microbial growth is less likely to promote selective pressure to develop antibacterial resistance35, C. xanthorhiza Roxb. ethanolic extract at 0-200 mg/mL were used to study the quorum quenching activity against P. aeruginosa.
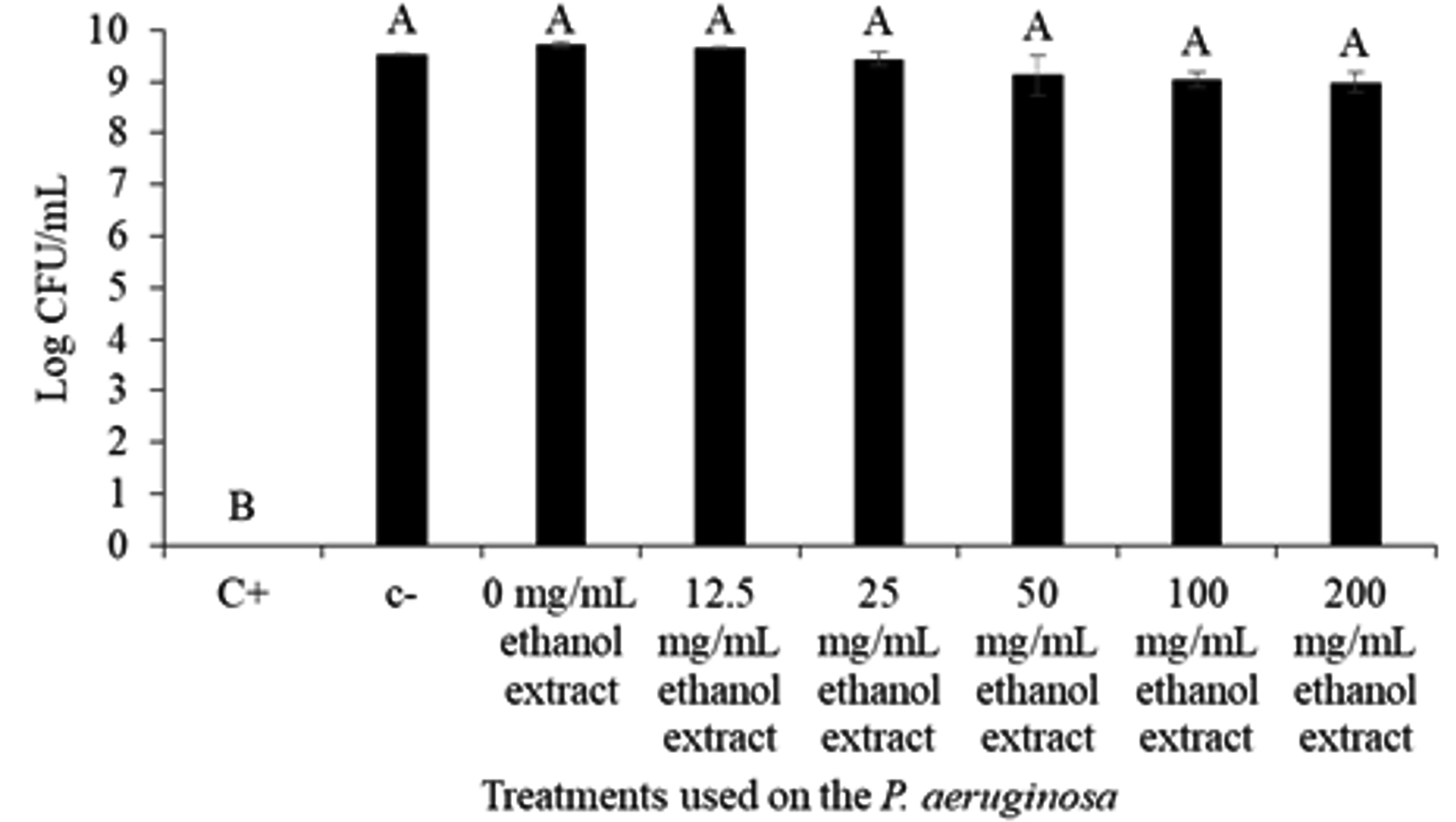 Fig. 1. Log10 CFU/mL of the P. aeruginosa ATCC 35554 growth against treatment with positive control (C+) 10 mg/mL Chlorhexidine, negative control (C-) DMSO, and C. xanthorrhiza Roxb. extract concentration (0-200 mg/mL). The alphabetical grouping represent the significance different between extract concentration (Those that share similar letter is not significant different).
Fig. 1. Log10 CFU/mL of the P. aeruginosa ATCC 35554 growth against treatment with positive control (C+) 10 mg/mL Chlorhexidine, negative control (C-) DMSO, and C. xanthorrhiza Roxb. extract concentration (0-200 mg/mL). The alphabetical grouping represent the significance different between extract concentration (Those that share similar letter is not significant different). Swarming assay
Inhibition on swarming motility was done by measuring the diameter of the swarming colonies in the presence of 0-200 mg/mL of C. xanthorrhiza Roxb. ethanolic extract. Fig. 2 display the decrement of P. aeruginosa swarming colony diameter. At 0 mg/mL, highest swarming colony was observed at 4.78 ± 0.29 cm and at 200 mg/mL, the extract managed to inhibit 72.12% of P. aeruginosa swarming with a diameter of 1.33 ± 0.12 cm. Crude extract of the spice clove (Syzygium aromaticum) has also been reported to exhibit reduction of P. aeruginosa PA0137,61. Since the swarming activity in P. aeruginosa is induced and regulated by the rhl system, presence of rhl inhibitor in the C. xanthorrhiza Roxb. ethanolic extact is associated with the reduction of P. aeruginosa swarming activity37.
 Fig. 2. Effect of the Curcuma xanthorrhiza Roxb. extract (0 – 200 mg/mL) on the P. aeruginosa swarming motility. The reduction of swarming diameter (cm) when added with C. xanthorrhiza Roxb. extract. The alphabetical grouping represent the significance different between extract concentration (Those that share similar letter is not significant different).
Fig. 2. Effect of the Curcuma xanthorrhiza Roxb. extract (0 – 200 mg/mL) on the P. aeruginosa swarming motility. The reduction of swarming diameter (cm) when added with C. xanthorrhiza Roxb. extract. The alphabetical grouping represent the significance different between extract concentration (Those that share similar letter is not significant different). Progression of swarming motility is hugely affected by the surfactant productions such as rhamnolipids (RLs) and 3-hydroxyalkanoic acids (HAAs)62-64. Such molecules affect swarming motility via inhibiting and promoting the tendril formation by displaying different diffusion kinetics on the agar64. Surfactant such as rhamnolipid depends on the activation of rhlA, rhlB and rhlC genes which are governed by the RhlR QS system and stimulated by the N-butyryl homoserine lactone induction65,66. Caiazza et al.67 have reported that mutation or inactivation on the rhlC gene that encodes the rhamnosyltransferases to initate the formation of monorhamnolipids can affect the inhibition on swarming motility.
In another study by Kim and Park68, ginger (Zingiber officinale) extract, instead of inhibiting the swarming activity of P. aeruginosa, the extract promoted its motility. Despite previous study by Rasmussen et al.69 have reported the extract quorum quenching ability such as on biofilm formation on P. aeruginosa PA14, no such inhibition was observed in Kim and Park’s study68. This phenomenon can be explained in the study by Caiazza et al.70 whereby the inverse regulation of P. aeruginosa swarming and biofilm activities is regulated by the flagella reversal and formation of Pel polysaccharides. Such mechanisms are crucial in the transition from swarming to biofilm formation by affecting the initial attachment between the bacterial and the substratum71.
O’may and Tufenkji72 reported that cranberry products which contains a condensed A-type of proanthocyanidins (PACs), hydrolysable tannin in pomegranate, catechins containing B-type PACs in the green tea extracts all managed to inhibit P. aeruginosa swarming, suggesting the quorum quenching of tannins. As swarming and biofilm require the QS system to effectively work, tannin compounds have been reported to be able to impede the mechanism73,74 as they are able to bind and precipitate various types of proteins75-77. Apart from tannins, phenols and phenolic compounds have also been reported to inhibit P. aeruginosa swarming motility35,78. Therefore, as the ethanol solvent of C. xanthorrhiza Roxb. exhibited the highest recovery of tannins45 and phenols and phenolic compounds27, the swarming inhibition of P. aeruginosa by the C. xanthhorhiza Roxb. extract could be due to its anti-QS properties.
Pyocyanin assay
In the pyocyanin inhibition assay, the extract was able to inhibit 84.30% of the pigment production by P. aeruginosa. The inhibition of pyocyanin pigment production was dependent on the extract concentration whereby at 0 and 200 mg/mL of the extract, 13.01 ± 1.37 mg/mL and 2.04 ± 0.59 mg/mL of pyocyanin concentration were produced (Fig. 3). During the extraction of pyocyanin, chloroform and 0.2 N HCl gave a blue and red colour, respectively. Pyoycanin colour and absorption spectrum have been reported to be pH sensitive and changes according to the exchange of electrons in the pigment79. For example, at a neutral pH, pycoyanin produces a strong blue colour, greenish blue at alkaline and red at an acidic pH79. Since of the three, the reduced form of pyocyanin have been reported to be unstable and reactive with molecular oxygen rapidly80,81, after extraction with chloroform, the pigment was re-extracted with 0.2 N HCl.
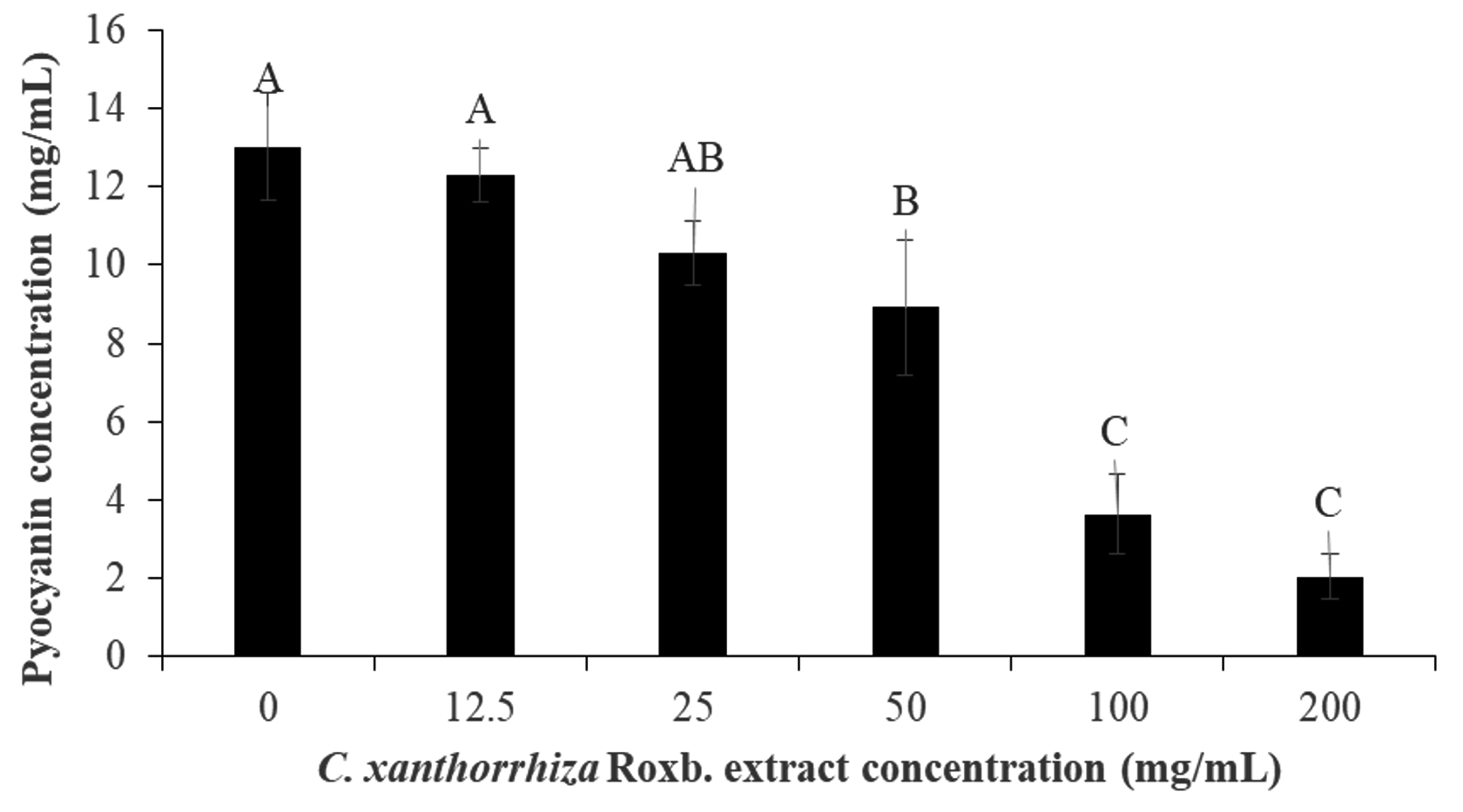 Fig. 3. Pyocyanin inhibition by the Curcuma xanthorrhiza Roxb. extract concentration (0-200 mg/mL). The pyoycanin concentration (mg/mL) after treated with Curcuma xanthorrhiza Roxb. extract concentration (0-200 mg/mL). The alphabetical grouping represent the significance different between extract concentration (Those that share similar letter is not significant different).
Fig. 3. Pyocyanin inhibition by the Curcuma xanthorrhiza Roxb. extract concentration (0-200 mg/mL). The pyoycanin concentration (mg/mL) after treated with Curcuma xanthorrhiza Roxb. extract concentration (0-200 mg/mL). The alphabetical grouping represent the significance different between extract concentration (Those that share similar letter is not significant different). Production of the pyocyanin pigment is commonly regulated by the Las, Rhl and PQS systems in which suggesting that P. aeruginosa pigment inhibition by C. xanthorrhiza Roxb. affect such systems by binding the autoinducers to its protein receptors nor interfering with pyocynin biosynthesis. In the study by Krishnan et al.37 suggested that the hexane, chloroform and methanol extract of S. aromatic or clove can affect P. aeruginosa production of swarming motility and pyocyanin pigment. Apart from chloroform, both hexane and methanolic clove extract exhibited reduction in pyocyanin production and this suggest that the inhibitions are done by the las and rhl inhibitors present in the extract37. Therefore, this could also suggest that the phytochemicals present in the C. xanthorrhiza Roxb. ethanolic extract might also consist of such inhibitors.
P aeruginosa pyocyanin pigment production has also been reported to be affected by the concentration of phenols and phenolic compounds. For example, phenolic compounds duch as vanilic acid, caffeic acid, cinnamic acid and ferulic acid prepared in ethanol/water mixtures were able to reduce approximately 9-21% pyoycanin production35. Such compounds were also reported to inhibit 50% of the biofilm formation and swarming motility by P. aeruginosa. Inhibitions of quorum sensing virulence activity and biofilm formation by phenolic compounds are also supported by various studies such as in the study by Borges et al.81 and the phenolic compounds present in Moringa oleifera82.
Meanwhile, Ab Halim et al.27 have made a study on the qualitative phytochemical screening of C. xanthorrhiza Roxb. extract to show presence of terpenoids, phenols, flavonoids, saponins, cardiac glycosids, alkaloids and coumarins. Futhermore, through the total phenol content (TPC) test on C. xanthorrhiza Roxb. ethanolic extract yielded a higher TPC value compared to its methanolic extract83,84. Phenols and its compounds have been known for its medicinal properties such as for skin infections and wound treatments85. Therefore, apart from its antibacterial, antifungal86 and other medicinal benefits, phenols and its compounds present in the C. xanthorrhiza Roxb. ethanolic extract might serves as a quorum quenching agent.
Biofilm assay
p aeruginosa biofilm formation was done colorimetrically to measure the inhibition via the absorbance at 495 nm when the XTT dye was added to the culture. Pyocyanin is involved in the formation of biofilm, therefore, in P. aeruginosa biofilm formation, green colour can be observed. Post incubation, the culture is washed with saline to remove the unattached culture to the wells and XTT dye was added. XTT dye is used as it contains tetrazolium salt that changes the colour to orange formazan in the presence of metabolic active cells in the biofilm87. In this assay, wells treated with 200 mg/mL of the extract showed the lightest orange colour, while well with 0 mg/mL extract showed the darkest orange colour. Fig. 4 shows the inhibition of the biofilm formation to be concentration dependent. Culture with no extract (0 mg/mL) exhibited the highest absorbance at 1.50 ± 0.048 but gradually decreases as the concentration of the extract increases. This is portrayed when the addition of 12.5 mg/mL of the extract managed to inhibit 24.07% of the biofilm formation while 200 mg/mL of the extract was able to inhibit it at 78.35% (1.14 ± 0.067 and 0.32 ± 0.035 of OD495 value, respectively).
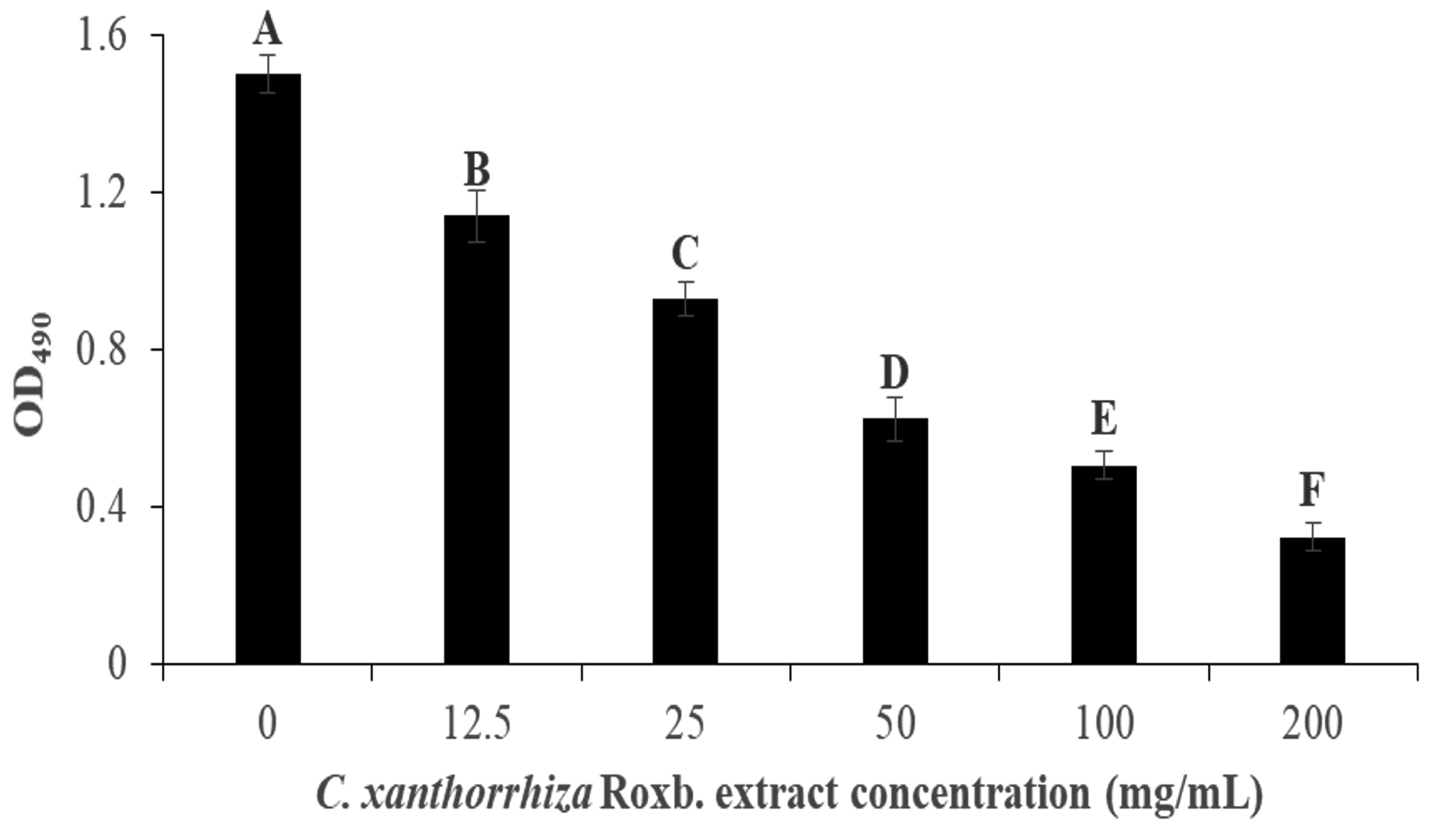 Fig. 4. The inhibition of the P. aeruginosa biofilm formation by C. xanthorrhiza Roxb. extract via absorbance at OD490. The alphabetical grouping represent the significance different between extract concentration (Those that share similar letter is not significant different).
Fig. 4. The inhibition of the P. aeruginosa biofilm formation by C. xanthorrhiza Roxb. extract via absorbance at OD490. The alphabetical grouping represent the significance different between extract concentration (Those that share similar letter is not significant different). Since the absorbance of the dye was shown to be decreasing as the extract concentration increases, this might suggest inhibition on biofilm formation in this assay due to increasing concentration of cell death in the biofilm88. In the study, up to 50% localized killing and lysis were observed in the biofilm center where the microcolonies are formed. There are several explanations that can attribute to biofilm cell death such as due reactive oxygen species (ROS) such as hydrogen peroxide (H2O2) and nitric oxide (NO) concentration accumulation in the quorum leading to localized lysis and biofilm cell deaths88,89.
Normally, NO concentration is regulated by the production of pyocyanin pigment via redox reaction90. Pyocyanin is also reported to facilitate biofilm formation via the release of extracellular DNA (eDNA) through H2O2 facilitated cell death91. In the early stages of biofilm development to form stable and mature biofilm, more than 50% of the biofilm matrixes are produced by eDNA92,93. eDNA plays a crucial factor by regulating the interconnection of P. aeruginosa cells57 such as promoting strong adherence towards the surface and providing a stable structure in the exopolysaccharides (EPS) of biofilm92,93 and involved in cell-to-cell communication92-94. Furthermore, eDNA also facilitates biofilm expansion via twitching motility by maintaining a coherent cell alignment95 and act as a nutrient source during starvation96,97.
Production of biofilm in P. aeruginosa has been reported to involve an inverse regulation between swarming versus biofilm and pyocyanin98. C-di-GMP, a compound that is found to be positively regulated in production of pyocyanin and biofilm was reported to be negatively regulated when swarming motility is high99-102. Such claim is also reported in the study by Kim and Park68 where by the ginger extract were able to reduce P. aeruginosa biofilm formation but increases the swarming activity.
Swarming and biofilm formation relationship involves the type of cell attachment to the surface. While swarming motility requires reversible and motile attachment, biofilm formation involves sessile, non-motile attachment. Reduction in biofilm formation occurs when the concentration of non-motile sessile cell attachment to the surface is decreasing98,103. The conversion between the two often regulated via expression of sadB and sadC genes that control the production of c-di-GMP concentration98,103. Since BifA is a C-di-GMP phosphodiesterase, interference on its activation can reduce c-di-GMP concentration thus reducing the signal C-di-GMP from being transmitted to Pel protein and CheIV chemotaxis-like cluster components104.
However, a contradiction was raised in the recent studies whereby both inhibitions on both quorum sensing regulation virulence were observed105,106. Both inhibition on swarming and biofilm formation can also be explained when the production of its signaling molecules are interrupted. This is supported by the study by Krishnan et al.37 whereby the presence of rhl inhibitor in clove extract were able to reduce swarming and biofilm production. Furthemore, C. xanthorrhiza Roxb. ethanolic extract was able to act as a quorum quenching agent by affecting the GacA/GacS system, a super QS regulator reported to aid the four QS systems in P. aeruginosa107. The super QS regulator can be affected by inactivating the free Rsma and increasing RetS activity, which will negatively regulate the production of AIs and repress biofilm formation, respectively108,109. Therefore, this could suggest that apart from inhibiting biofilm maturation, swarming motility and pyocyanin pigment production by affecting the concentration of c-di GMP associated mechanisms, the C. xanthorrhiza Roxb. extract also interfered with the production of the QS autoinducers signaling molecules and systems, thus affecting the activity of swarming, pyoycanin and biofilm formation.
In conclusion, as the extract concentration chose did not exhibited or only slightly show the antimicrobial activity against P. aeruginosa bacterial growth, the inhibition on the quorum sensing mediated virulence and biofilm formation could be due to the intrinsic quorum quenching activities by the extract. The ethanolic extract of C. xanthorrhiza Roxb. effect on the P. aeruginosa quorum sensing is concentration dependent whereby at 200 mg/mL, the extract were able to inhibit 72.12% of P. aeruginosa swarming, 84.30% of pyocyanin production and 78.35% of the biofilm formation. These highlight the extracts’ potential as a good anti-quorum sensing agent.
Acknowledgements
The authors would like to acknowledge financial support from Geran Putra UPM with number GP-IBT/2013/9410800 to Yaya Rukayadi.
Conflicts Of Interest
The authors declares that there is no conflict of interest.
Authors’ Contribution
AFO substantially contributed in investigation and writing original draft. YR substantially contributed in conception, design of the work and interpreted microbiological data. YR and SR did the draft editing and reviewed the manuscript.
Funding
This research was founded by Universiti Putra Malaysia under Geran Putra UPM with number GP-IBT/2013/9410800 to Yaya Rukayadi.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Fukuda K. Food safety in a globalized world. Bull. World Health Organ, 2015; 93: 212. Crossref
- Gustavsson J., Cederberg C., Sonesson U., Otterdijk R.V., Meybeck, A. Global Food Losses and Food Waste: Extent, Causes and Prevention, 2011. Rome: Food and Agriculture Organization of the United Nations, Natural Resources Management and Environment Department, 2011.
- Adams MS., Moss MO. Food Microbiology. 2nd Ed. Cambridge, UK: The Royal Society of Chemistry, 2000.
- Mortality and Global Health estimate. https://www.who.int/gho/en/, Accessed 7 July 2018.
- Scallan E., Hoekstra RM., Angulo FJ., Tauxe RV., Widdowson M.A., Roy S.L., Griffin P.M. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis., 2011; 17(1): 7–15.
Crossref - Teisl MF., Roe BE. Consumer willingness-to-pay to reduce the probability of retail foodborne pathogen contamination. Food Policy, 2010; 35: 521–530.
Crossref - Abdul-Mutalib NA, Syafinaz AN., Sakai K., Shirai Y. An overview of foodborne illness and food safety in Malaysia. Int. Food Res. J., 2015; 22(3): 896–901.
- Soon JM., Singh H., Baines R. Foodborne diseases in Malaysia: A review. Food Control, 2011; 22: 823–830.
Crossref - Levinson W. Review of Medical Microbiology and Immunology, pp. 142-159. 13th Ed. The McGraw Hill Education, New York, 2014.
- Sigma Aldrich. Mechanism of Action – Antibiotics. https://www.sigmaalrich.com, Accessed 20 May, 2018.
- Bhunia AK. Introduction of Foodborne Pathogens, pp. 1-16. In Foodborne Microbial Pathogens. 1st Ed. Springer, New York, 2008.
Crossref - Dong Y., Wang L., Zhang LH. Quorum-quenching microbial infections: Mechanisms and implications. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 2007; 362(1483): 1201–1211.
Crossref - Zhang LH., Dong YH. Quorum sensing and signal interference: Diverse implications. Mol. Microbiol., 2004; 53(6): 1563–1571.
Crossref - Ahmad A., Viljoen AM., Chenia HY. The impact of plant volatiles on bacterial quorum sensing. Lett. Appl. Microbiol., 2015: 60(1): 8–19.
Crossref - Skandamis PN., Nychas GJE. Quorum sensing in the context of food microbiology. Appl. Environ. Microbiol., 2012; 78(16): 5473–5482.
Crossref - Szabo M., Varga GZ., Hohmann J., Schelz Z., Szegedi E., Amaral L., Molnar J. Inhibition of quorum-sensing signals by essential oils. Phytother. Res., 2010; 24(5): 782–786.
Crossref - Bai AJ., Rai VR. Bacterial Quorum Sensing and Food Industry. Compr. Rev. FoodSci. Food Saf., 2011; 10(3): 183–193.
Crossref - Hossain Z. Bacteria, pp. 490–491. In Motarjemi Y, Moy G, and Ewen CDT (eds.), Encyclopedia of Food Safety, 1st Ed. Academic Press, Cambridge, Massachusetts, 2013.
Crossref - Lee J., Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein and Cell, 2014; 6(1): 26–41.
Crossref - Lew KF., Goh GL., Son R., Rukayadi Y. Effect of Javanese turmeric (Curcuma xanthorrhiza Roxb.) extract on natural microflora of oyster mushroom (Pleurotus sajur-caju) and its sensory acceptability. Int. Food Res. J., 2015; 22(6), 2446–2451.
- Musfiroh I., Muchtaridi M., Muhtadi A., Diantini A., Hasanah AN., Udin LZ., Ibrahim S. Cytotoxicity studies of xanthorrhizol and its mechanism using molecular docking simulation and pharmacophore modelling. J. Appl. Pharm. Sci., 2013; 3(6): 7-15.
- Lim TK. Curcuma zanthorrhiza. pp. 384. In Edible Medicinal and Non-Medicinal Plants. Cham: Springer International Publishing, New York, 2016.
Crossref - Park JH., Park KK., Kim MJ., Hwang JK., Park SK., Chung WY. Cancer chemoprotective effects of Curcuma xanthorrhiza. Phytother. Res., 2008; 22(5): 695–698.
Crossref - Jantan I., Saputri FC., Qaisar MN., Buang F. Correlation between chemical composition of Curcuma domestica and Curcuma xanthorrhiza and their antioxidant effect on human low-density lipoprotein oxidation. Evid. Based Complement. Alternat. Med., 2012; 438356.
Crossref - Sylvester WS., Son R., Lew KF., Rukayadi Y. Antibacterial activity of Java turmeric (Curcuma xanthorrhiza Roxb.) extract against Klebsiella pneumoniae isolated from several vegetables. Int. Food Res. J., 2015; 22(5): 1770–1776.
- Casilag F., Lorenz A., Krueger J., Klawonn F., Weiss S., Haussler S. LasB elastase of Pseudomonas aeruginosa acts in concert with alkaline protease AprA to prevent flagellin-mediated immune recognition. Infect. Immun., 2015; 84(1): 162–171.
Crossref - Ab Halim MR., Mahmud MZT., Mahmud R., Ismail S. Standardization and phytochemical studies of Curcuma xanthorrhiza Roxb. Int. J. Pharm. Pharm. Sci., 2012; 4(3): 606–610.
- Soniya M., Kuberan T., Anitha S., Sankareswari P. In vitro antibacterial activity of plant extracts against Gram positive and Gram negative pathogenic bacteria. I. J. Microbiol. Immun. Res., 2013; 2(1): 1–5.
- Naz R., Ayub H., Nawaz S., Islam ZU., Yasmin T., Bano A., Roberts TH. Antimicrobial activity, toxicity and anti- inflammatory potential of methanolic extracts of four ethnomedicinal plant species from Punjab, Pakistan. BMC Complement. Altern Med., 2017; 302: 1-13.
Crossref - Clinical and Laboratory Standards Institute (CLSI). 2005. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard, pp. M2-A8. 8th Ed. Villanova, PA.
- Hazan R., Que YA., Maura D., Rahme LG. A method for high throughput determination of viable bacteria cell counts in 96-well plates. BMC Microbiol., 2012; 12: 259.
Crossref - Rashid MH., Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Nat. Acad. Sci., 2000; 97(9): 4885–4890.
Crossref - King M., Guragain M., Sarkisova S., Patrauchan M. Pyocyanin extraction and quantitative analysis in swarming Pseudomonas aeruginosa. Bio-Protocol,.
Crossref - Karpagam S., Sudhakar T., Lakshmipathy M. Microbicidal response of pyocyanin produced by Pseudomonas aeruginosa toward clinical isolates of fungi. Int. J. Pharm. Pharm. Sci., 2013; 5(3): 870–873.
- Ugurlu A., Yagci AK., Ulusoy S., Aksu B., Bosgelmez-Tinaz G. Phenolic compounds affect production of pyocyanin, swarming motility and biofilm formation of Pseudomonas aeruginosa. Asian Pac. J. Tropic. Biomedic, 2016; 6(8): 698-701.
Crossref - Ozcan D., Kahraman H. Pyocyanin production in the presence of calcium ion in Pseudomonas aeruginosa and recombinant bacteria. Turkish J. Sci. Technol., 2015; 10(1): 13–19.
- Krishnan T., Yin WF., Chan KG. Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa PAO1 by ayurveda spice clove (Syzygium aromaticum) bud extract. Sensors, 2012; 12(4): 4016–4030.
Crossref - Datta S., Debanjan Jana B., Tilak Raj Maity B., Aveek Samanta B., Rajarshi Banerjee B. Piper betle leaf extract affects the quorum sensing and hence virulence of Pseudomonas aeruginosa PAO1. Biotech, 2016; 6:18.
Crossref - Chong YM., Yin W., Ho CY., Mustafa MR., Hadi A., Hamid A., Awang K., Narrima P., Koh CL., Appleton DR., Chan KG. Malabaricone C from Myristica cinnamomea exhibits anti-quorum sensing activity. J. Nat. Prod., 2011; 74 (10): 2261-2264.
Crossref - Sarkisova S., Patrauchan MA., Berglund D., Nivens DE., Franklin MJ. Calcium-induced virulence factors associated with the extracellular matrix of mucoid Pseudomonas aeruginosa biofilms. Society, 2005; 187(13): 4327–4337.
Crossref - Essar DW., Eberly L., Hadero A., Crawford IP. Identification and characterization of genes for a 2nd Anthranilate Synthase in Pseudomonas aeruginosa: Interchangeability of the 2 anthranilate synthases and evolutionary implications. J. Bacteriol., 1990; 172(2): 884–900.
Crossref - Varposhti M., Abdi Ali A., Mohammadi P., Saboora A. Effects of extracts and an essential oil from some medicinal plants against biofilm formation of Pseudomonas aeruginosa. J. Med. Microbiol. Infect. Dis., 2013; 1(1): 36–40.
- Azwanida NN. A review on the extraction methods use in medicinal plants, principle, strength and limitation. Med. Aromat. Plants, 2015; 4: 196.
- Do QD., Angkawijaya AE., Tran-Nguyen PL., Huynh LH., Soetaredjo FE., Ismadji S., Ju YH. Effect of extraction solvent on total phenol content, total flavonoid content and antioxidant activity of Limnophila aromatica. J. Food Drug Anal., 2014; 22(3): 296–302.
Crossref - Aida W. Effect of ethanol concentration, extraction time and extraction temperature on the recovery of phenolic compounds and antioxidant capacity of Centella asiatica extracts. Int. Food Res. J., 2011; 18: 571–578.
- Dai J., Mumper R.J. Plant phenolics: Extraction, analysis and their antioxidant and anticancer properties. Molecules, 2010; 15: 7313-7352.
Crossref - Whitcombe T. Why is ethanol the only relatively safe alcohol for human consumption?. MadSci Network: Biochemistry, 2000. http://www.madsci.org/posts/archives/2000-11/973793825.Bc.r.html, Accessed 17 July 2018.
- Saifudin A., Rahayu HV., Teruna HY. “Standarisasi bahan obat alam”. Pp. 16. Graha Ilmu. Yogyakarta, 2011.
- Tarigan J., Zuhra CF., Sihotang H. Skrining fitokimia tumbuhan yang digunakan oleh pedagang jamu gendong untuk merawat kulit wajah di Kecamatan Medan Baru. J. Biol. Sumatera, 2008; 1(3): 1–6.
- Diastuti H., Syah YM., Juliawaty LD., Singgih M. Antibacterial Curcuma xanthorrhiza extract and fractions. J. Math. Fund. Sci., 2014; 46(3): 224–234.
Crossref - Harborne J. Phytochemical methods. A guide to modern techniques of plants analysis. 2nd Ed. Chapman and Hall, London, 1996.
- Mangunwardoyo W., Deasywaty, Usia T. Antimicrobial and identification of active compound Curcuma xanthorrhiza Roxb. Int. J. Basic Appl. Sci. IJBAS-IJENS, 2012; 12(1): 69–78.
- Selim S., Hassan S., Al Soumaa K., EL-Anzy S. Prevalence, antibiotic resistance and in vitro activity of yogurt against some Gram negative pathogenic bacteria isolated from Arar Hospital, KSA. Life Sci. J., 2013; 10: 1450–1456.
- Hanouda T., Baker JR. Antimicrobial mechanism of action of surfactant lipid preparation in enteric Gram negative bacilli. J. Appl. Microbiol., 2000; 89: 397–403.
Crossref - Hidayathulla S., Keshava CK., Chandrashekar KR. Phytochemical evaluation and antibacterial activity of Pterospermum diversifolium Blume. Int. J. Pharm. Sci., 2011; 3(2): 165–167.
- Al-Rubiay KK., Jaber NN., Al Mhaawe BH., Alrubaay LK. Antimicrobial of henna extract. Oman Med. J., 2008; 23: 4 – 8.
Crossref - Nohynek LJ., Alakomi A., Kankonen MP., Heinonen M., Helander I.M., Oksman- Caldentey KM., Puupponen-Pimia RH. Berry phenolics: Antimicrobial properties and mechanisms of action against severe human pathogens. Nutr. Cancer, 2006; 54(1): 18–32.
Crossref - Taylor PK., Yeung ATY., Hancock REW. Antibiotic resistance in Pseudomonas aeruginosa biofilms: Towards the development of novel anti-biofilm therapies. J. Biotechnol., 2014; 191: 121–130.
Crossref - Hertiani T., Palupi SI., Sanliferianti., Nurwindasari D.H. In vitro test on antimicrobial potency against Staphyloccocus aureus, Escherichia coli, Shigella dysentriaea and Candida albicans of some herbs traditionally used cure infection diseases. Pharmacon, 2003; 4(2): 89–95.
- Hancock REW., and Bell A. Antibiotic uptake into Gram-negative bacteria. Eur. J. Clin. Microbiol. Infect. Dis., 1988; 7: 713–720.
Crossref - Khan MSA., Zahin M., Hasan S., Husain FM., Ahmad I. Inhibition of quorum sensing regulated bacterial functions by plant essential oils with special reference to clove oil. Lett. Appl. Microbiol., 2009; 49(3): 354–360.
Crossref - Kצhler T., Curty LK., Barja F., van Delden C., Pechטre JC. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol., 2000; 182(21): 5990–6.
Crossref - Davey ME., Caiazza NC., O’Toole GA. Rhamnolipid surfactantproduction affects biofilm architechture in Pseudomonas aeruginosa PA01. J. Bacteriol., 2003; 185(3): 1027-1036.
Crossref - Nogales J., Dominiguez-Ferreras A., Amaya-Gomez CV., van Dillewijn P., Cuellar V., Sanjuan J., Olivares J., Soto M.J. Transcriptome profiling of a Sinorhizobium meliloti fadD mutant reveals the role of rhizobactin 1021 biosynthesis and regulation genes in the control of swarming. BMC Genomics, 2010; 11: 157.
Crossref - Rahim R., Ochsner UA., Olvera C., Graninger M., Messner P., Lam J.S., Soberףn-Chavez G. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes Rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol., 2001; 40(3): 708–18.
Crossref - Ochsner UA., Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Nat. Acad. Sci., 1995; 92: 6424–6428.
https://doi.org/10.1073/pnas.92.14.6424. - Caiazza NC., Shanks RMQ., O’toole GA. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol., 2005; 187(21): 7351–7361.
https://doi.org/10.1128/JB.187.21.7351-7361.2005. - Kim HS., Park HD. Ginger extract inhibits biofilm formation by Pseudomonas aeruginosa PA14. PLoS ONE, 2013; 8: 9.
https://doi.org/10.1371/journal.pone.0076106. - Rasmussen TB., Bjarnsholt T., Skindersoe ME., Hentzer M., Kristoffersen P., Kצte M., Givskov M. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol., 2005; 187(5): 1799–814.
https://doi.org/10.1128/JB.187.5.1799-1814.2005. - Caiazza NC., Merritt JH., Brothers KM., O’Toole GA. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol., 2007; 189(9): 3603–12.
https://doi.org/10.1128/JB.01685-06. - Pratt LA., Kolter R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol., 1998; 30(2): 285–293.
https://doi.org/10.1046/j.1365-2958.1998.01061.x. - O’May C., Tufenkji N. The swarming motility of Pseudomonas aeruginosa is blocked by Cranberry proanthocyanidins and other tannin-containing materials. Appl. Environ. Microbiol., 2011; 77(9): 3061–3067.
https://doi.org/10.1128/AEM.02677-10. - Vattem DA., Mihalik K., Crixell SH., McLean RJC. Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia, 2007; 78(4): 302–310.
https://doi.org/10.1016/j.fitote.2007.03.009. - Huber B., Eberl L., Feucht W., Polster J. Influence of polyphenols on bacterial biofilm formation and quorum-sensing. Z. Naturforsch, 2003; 58: 879–884.
https://doi.org/10.1515/znc-2003-11-1224. - Serrano J., Puupponen-Pimiה R., Dauer A., Aura AM., Saura-Calixto F. Tannins: Current knowledge of food sources, intake, bioavailability and biological effects. Mol. Nut. Food Res., 2009; 53(S2): S310–S329.
https://doi.org/10.1002/mnfr.200900039. - Haslam E. Natural polyphenols (vegetable tannins) as drugs: Possible modes of action. J. Nat. Prod., 1996; 59(2): 205–15.
https://doi.org/10.1021/np960040+. - Hagerman AE., Butler LG. The specificity of proanthocyanidin-protein interactions. The J. Biol. Chem., 1981; 256(9): 4494–7.
- Borges A., Saavedra MJ., Simץes M. The activity of ferulic and gallic acids in biofilm prevention and control of pathogenic bacteria. Biofouling, 2012; 28 (7): 755–767.
Crossref - Reszka KJ., O’Malley Y., McCormick ML., Denning GM., and Britigan BE. Oxidation of pyocyanin, a cytotoxic product from Pseudomonas aeruginosa, by microperoxidase 11 and hydrogen peroxide. Free Radical Biology and Medicine, 2004; 36(11): 1448–1459.
Crossref - Prijaya P., Philip R., Singh ISB. Cloning and overexpression of Phz genes encoding phenazine biosynthetic pathway for the enhanced production of pyocyanin in Pseudomonas aeruginosa MCCB117. Cochin University of Science and Technology, Shodhganga. India, 2013.
- Borges A., Serra S., Cristina Abreu A., Saavedra MJ., Salgado A., Simץes M. Evaluation of the effects of selected phytochemicals on quorum sensing inhibition and in vitro cytotoxicity. Biofouling, 2014; 30(2): 183–195.
Crossref - Karatuna O., Yagci A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin. Microbiol. Infect., 2010; 16(12): 1770–1775.
Crossref - Mustafa RA., Abdul Hamid A., Mohamed S., Bakar FA. Total phenolic compounds, flavonoids, and radical scavenging activity of 21 selected tropical plants. J. Food Sci., 2010; 75: C28-35.
Crossref - Moure A., Cruz JM., Franco D., Manuel Domםnguez J., Sineiro J., Domםnguez H., Nתסez M.J., Carlos Parajף J. Natural antioxidants from residual sources. Food Chem., 2001; 72: 145–171.
Crossref - Okwu D.E. Evaluation of the chemical composition of indigenous. Spices and flavouring agents. Global J. Pure Appl.Sci., 2001;7: 455–459.
Crossref - Kim JE., Kim HE., Hwang JK., Lee HJ., Kwon HK., Kim BI. Antibacterial characteristics of Curcuma xanthorrhiza extract on Streptococcus mutans biofilm. J. Microbiol., 2008; 46: 228–232.
Crossref - Tunney MM., Ramage G., Field TR., Moriarty TF., Storey DG. Rapid colorimetric assay for antimicrobial susceptibility testing of Pseudomonas aeruginosa. Antimicrob. Agents Chemother., 2004; 48(5): 1879-1881.
Crossref - Webb JS., Thompson LS., James S., Charlton T., Tolker-Nielsen T., Koch B., Givskov M., Kjelleberg S. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol., 2003; 185(15): 4585-4592.
Crossref - Barraud N., Hassett DJ., Hwang SH., Rice SA., Kjelleberg S., Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol., 2006; 188(21): 7344–7353.
Crossref - Samanta S., Thavasi R., Jayalakshmi S. Phenazine pigments from Pseudomonas aeruginosa and their application as antibacterial agent and food colourants. Res. J. Microbiol., 2008; 3(3): 122–128.
Crossref - Das T., Manefield M. Pyocyanin Promotes Extracellular DNA Release in Pseudomonas aeruginosa. PLOS ONE, 2012; 7(10): e4671B.
Crossref - Allesen-Holm M., Barken KB., Yang L., Klausen M., Webb JS., Kjelleberg S., Tolker-Nielsen TA. Characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol., 2006; 59: 1114–1128.
Crossref - Whitchurch CB., Tolker-Nielsen T., Ragas PC., Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science, 2002; 295(5559): 1487.
Crossref - Flemming HC., Wingender J. The biofilm matrix. Nat. Rev. Microbiol., 2010; 8(9): 623–633.
Crossref - Gloag ES., Turnbull L., Huang A., Vallotton P., Wang H., Nolan LM., Whitchurch CB. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc. Nat. Acad. Sci., 2013; 110(28): 11541–11546.
Crossref - Mulcahy H., Charron-Mazenod L., Lewenza S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol., 2010; 12(6): 1621–9.
Crossref - Finkel SE., Kolter R. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol., 2001; 183(21): 6288–93.
Crossref - Merighi M., Lee VT., Hyodo M., Hayakawa Y., Lory S. The second messenger bis-(3’-5’)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol., 2007; 65(4): 876–895.
Crossref - Merritt JH., Brothers KM., Kuchma SL., O ’toole GA. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol., 2007; 189(22): 8154–8164.
Crossref - Rצmling U., Gomelsky M., Galperin MY. C-di-GMP: The dawning of a novel bacterial signalling system. Mol. Microbiol., 2005; 57(3): 629–639.
Crossref - D’Argenio DA., Miller SI. Cyclic di-GMP as a bacterial second messenger. Microbiology, 2004; 150(8): 2497–2502.
Crossref - Jenal U. Cyclic di-guanosine-monophosphate comes of age: A novel secondary messenger involved in modulating cell surface structures in bacteria. Curr. Opin. Microbiol., 2004; 7: 185–191.
Crossref - O’toole GA. How Pseudomonas aeruginosa regulates surface behaviors. Microbe, 2008; 3(2): 65–71.
Crossref - Verstraeten N., Braeken K., Debkumari B., Fauvart M., Fransaer J., Vermant J., Michiels J. Living on a surface: Swarming and biofilm formation. Trends Microbiol., 2008; 16(10): 496–506.
Crossref - Vasavi H., Arun A., Rekha P. Anti-quorum sensing activity of flavonoid- rich fraction from Centella asiatica L. against Pseudomonas aeruginosa PAO1. J. Microbiol. Immun. Infect., 2016; 49: 8–15.
Crossref - Issac A., Sybiya VP., Palani A., Khadar SM., Shunmugiah KP., Arumugam VR. Antibiofilm and quorum sensing inhibitory potential of Cuminum cyminum and its secondary metabolite methyl-eugenol against Gram negative bacterial pathogens. Food Res. Int., 2012; 45: 85–92.
Crossref - Parkins MD., Ceri H., Storey DG. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol., 2001; 40(5): 1215–26.
Crossref - Kay E., Humair B., Dיnervaud V., Riedel K., Spahr S., Eberl L., Haas D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol., 2006; 188(16): 6026–33.
Crossref - Reimmann C., Beyeler M., Latifi A., Winteler H., Foglino M., Lazdunski A., Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol., 1997; 24(2): 309–19.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


